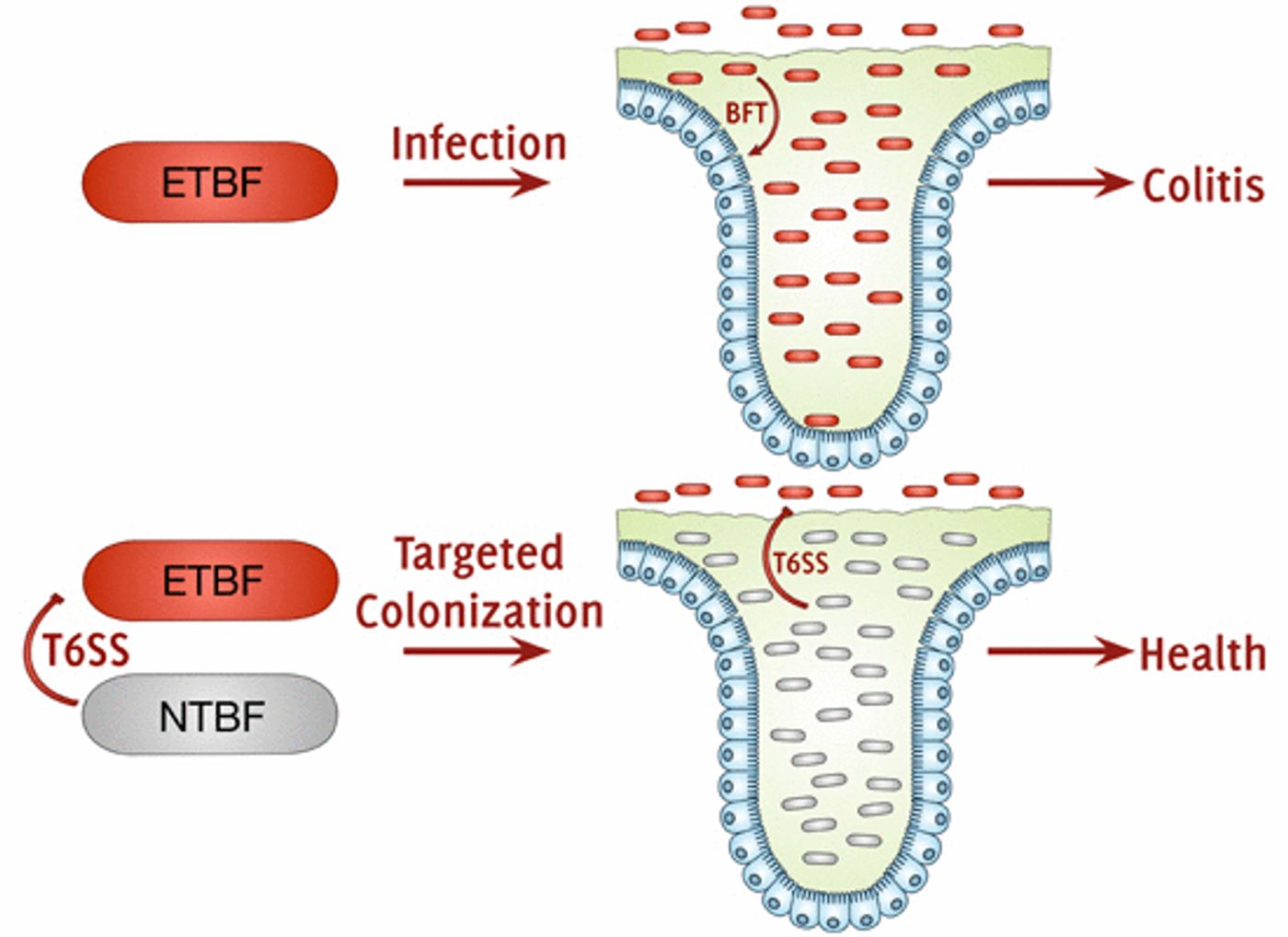
Commensal bacteria, the normal flora in the digestive tract, have a multitude of functions and are critical to health. One of those functions is keeping harmful or pathogenic bacteria out of our guts. Enterotoxigenic Bacteroides fragilis (ETBF) is one such bacterium.
A new study has used “specific-pathogen free” (SFP) mice to investigate the gut microbiome. While these are not truly germ-free, sterile mice, they get tested so they can be confirmed to be free of a specific list of pathogens. Those ‘pathogens’ can be disease-causing microbes, or they can just be commensal organisms.
The paper, published in EMBO Reports, shows that an effector can exclude the ETBF bacterial strain from the gut. Interestingly, a non-toxigenic B. fragilis (NTBF) strain secretes that effector, which thus indicates how strain-specific competition may work to mold the composition of gut microflora communities.
The gut microbiome of humans carries B. fragilis populations that are known to be primarily made up of either ETBF (toxic) or NTBF (non-toxic) strains. That’s a feature that indicated to the researchers that competition could be taking place and may aid in the study of the mechanism of pathogen exclusion.
The type VI secretion system (T6SS), explained in the video above, is a one step mechanism by which bacteria inject effector proteins and virulence factors like proteins or enzymes, from the bacterial cell and directly into a target cell.
It’s been previously demonstrated that T6SSs in B. fragilis manage competition between strains. The researchers compared exclusion of ETBF in strains of NTBF that were normal, or T6SS-deficient (the gene coding for an essential T6SS component tssC was deleted), or T6SS-complemented (tssC was removed from the genome but expressed on a plasmid).
By co-colonizing SPF mice, it was observed that normal NTBF and T6SS-complemented NTBF displayed strain dominance over ETBF. That dominance was abolished, however, in mice that were co-colonized with NTBF that was T6SS-deficient. It was confirmed that the phenotype of T6SS deficiency was caused by a failure to exclude toxic ETBF, and not from a loss NTBF, after monocolonization by NTBF was not different between normal and T6SS-deficient strains.
The scientists also wanted to see if disease burden was reduced after EPTF was excluded, so they assayed for the toxin it produces. They determined that interstrain competition allowed normal NTBF to provide total protection from ETBF-induced colitis, while T6SS-deficient NTBF only gave partial protection.
Using mass spectrometry to identify the effector, the T6SS secretome was characterized and Bte2 (B. fragilis T6S effector 2) was identified as the most abundant effector. In addition, analysis of an NTBF strain a deficient in Bte2 confirmed that it’s an effector that mediates exclusion of ETBF, and by consequence, lessens the disease burden in the mouse gut.
Consideration of all the data shows that Bte2 secretion by NTBF T6SS shapes the composition of the microbiome and reduces disease burden by targeting EBTF.
Sources:
Nature,
EMBO Reports