An Iterative Approach to Structure-based in silico to in vivo Development of Combinatorial Anti-SARS-CoV-2 Therapy
-
Garth D. Ehrlich, PhD, FAAAS
Professor of Microbiology & Immunology, Professor of Otolarynology-Head and Neck Surgery, Drexel University College of MedicineBIOGRAPHY
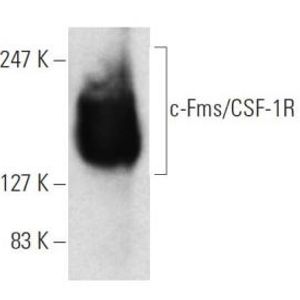
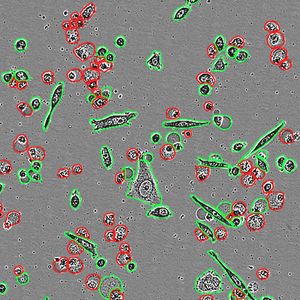
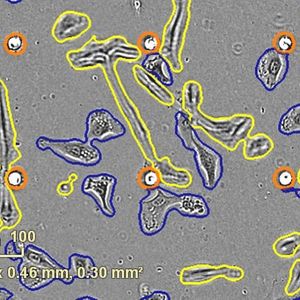
This drug development program is designed to create a family of broad-spectrum, pan-coronaviral drugs that respectively inhibit multiple key enzymes required for viral replication. By targeting multiple gene products simultaneously we hope to eliminate/minimize the development of treatment-resistant viral strains as was successfully done when HAART (highly active anti-retroviral therapy) was introduced for HIV-1 disease. We employ a pipeline-based drug development process that includes multiple feedback loops between and among both in silico and laboratory-based processes for the generation of highly effective and mutation-resistant pharmaceuticals. As part of our process to minimize the risk of the evolution of escape mutants, we begin our drug design using a suite of artificial intelligence-based machine-learning algorithms that are proven to: 1) predict future evolution (mutations) of the target viral enzymes in silico – these sequences are added to the extant set of all protein structure files from the -coronaviruses that correspond to each of our target enzymes; 2) perform protein structure overlays of all available structures for each target enzyme to determine the most highly conserved active site morphologies; 3) prepare a 3-D grid of the conserved (consensus) active site for each target enzyme; 4) using a 3-step in silico funnel process, with increased-stringency at each step, run each consensus target active-site against multiple large druggable compound libraries, including all FDA-approved drugs, to obtain ‘hit’ compounds. These ‘hit’ compounds are then simultaneously moved into: a) our in silico molecular dynamics pipeline to identify and characterize the active-site interacting groups (molecular domains) which will be used as one of the inputs into our structure-activity relationship (SAR) modeling to rationally build derivative compounds with greater affinities; and b) our three-phase laboratory biophysical-biochemical pipeline consisting of (i) surface plasmon resonance for protein binding kinetics, (ii) functional enzyme inhibition studies, and (iii) isothermal titration calorimetry and microscale thermophoresis to obtain precise stoichiometry and binding affinities under multiple liquid-phase parameters. Hit compounds meeting both preset biophysical and biochemical criteria will be moved as “leads” into the antiviral testing arm of the pipeline. Those compounds showing promise, but without high enough binding scores or IC50 values will be moved in the SAR pipeline for the development of rationally-designed derivatives. Results from the SAR studies will be screened for their ease/predicted yields of synthesis, and then process-specific software will be used to augment protocols for their organic syntheses. Lead compounds will be evaluated individually, and as cocktails of two and three compounds targeting different enzymes, for: 1) human cell toxicity; 2) SARS-CoV-2 virological studies using Calu-3 cells and human airway epithelial cells; and 3) in a physiologically relevant lung-on-a-chip system. Emergence of viral escape mutants in response to lead compounds will be sequenced and inform the synthesis of escape-resistant antiviral drugs. Finally, the prophylactic/therapeutic in vivo antiviral efficacy of lead compounds will be determined in animal models of coronavirus infection and pathogenesis.
Learning Objectives:
1. Learn applications of deep-learning algorithms and machine-learning techniques to help with the design of escape-resistant drugs for infectious agents
2. Learn targeting strategies to minimize the evolution of escape mutants
An Iterative Approach to Structure-based in silico to in vivo Development of Combinatorial Anti-SARS-CoV-2 Therapy
Please update your information
Certificate of Attendance
DOWNLOAD CERTIFICATE
Finish Registering
-
APR 30, 2024Immuno-Oncology Virtual Event Series 2024
-
MAY 07, 20243rd International Biosecurity Virtual Symposium
-
SEP 03, 2024Microbiology Week Virtual Event Series 2024
- See More
-
APR 17, 2024
- See More