Visualizing the Organization of Human Cells With Machine Learning
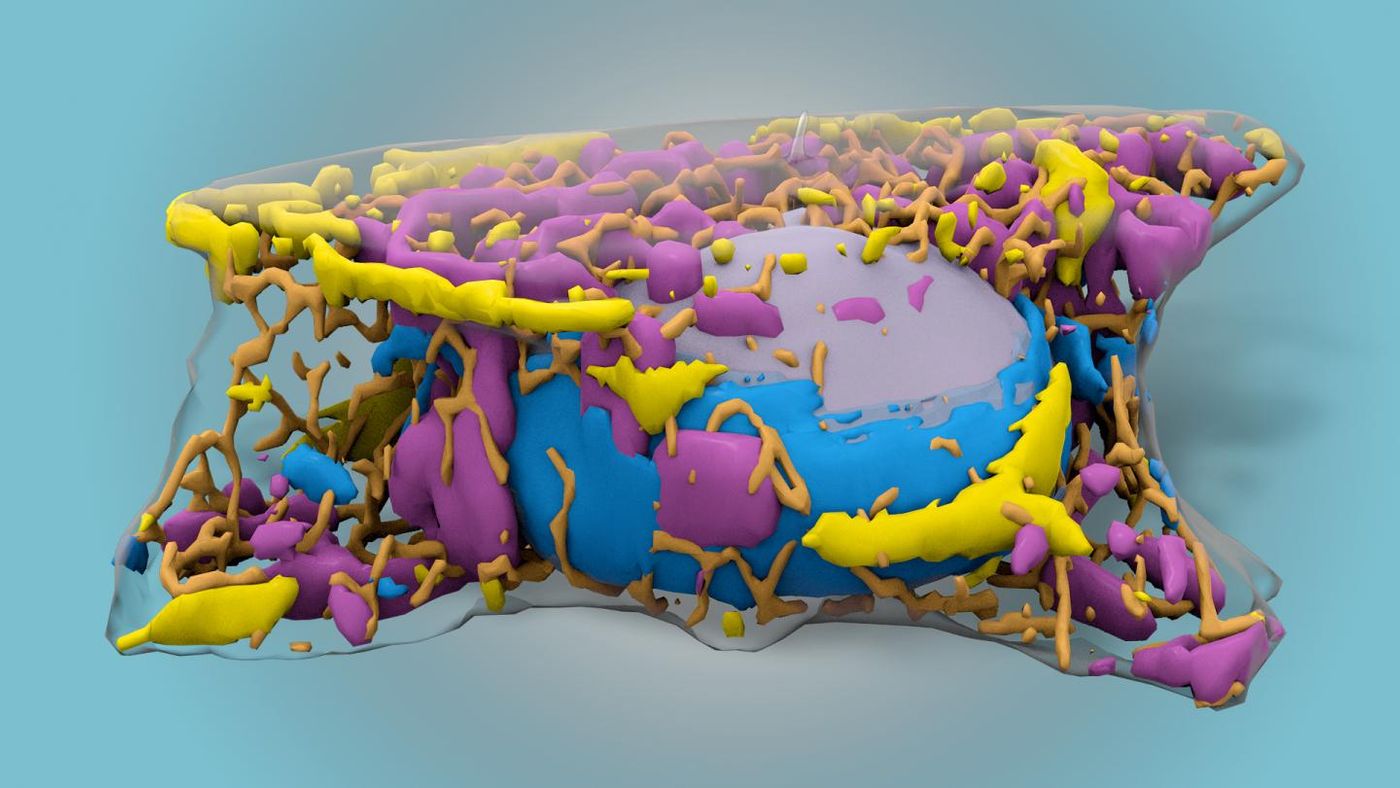
To see structures within cells, scientists have to use labelling tools to tag the structures or proteins they’re investigating. While that is a precise method, there are many limitations; it only labels a few things at a time, and it usually requires the cell to be fixed, so it’s no longer alive. Now researchers at the Allen Institute are taking us closer to being able to visualize more of the roughly 20,000 proteins that make up the structures and carry out the operations of a cell. With machine learning, computers were trained to identify subcellular structures with only black and white images taken with standard, inexpensive brightfield microscopy. Reporting in Nature Methods, the team describes how more cell structures can now be visualized at once.
"This technology lets us view a larger set of those structures than was possible before," said Greg Johnson, Ph.D., Scientist at the Allen Institute for Cell Science, a division of the Allen Institute, and senior author on the study. "This means that we can explore the organization of the cell in ways that nobody has been able to do, especially in live cells."
Aside from being used primarily on fixed cells, fluorescently-tagged cells require fluorescence microscopes for evaluation, which can be very expensive. The tags also fade over time, or when intense light from those microscopes is applied. This system has the potential to dramatically lower imaging costs associated with studying subcellular dynamics.
This predictive technique may also provide valuable insight into cells that are impacted by disease, noted Rick Horwitz, Ph.D., Executive Director of the Allen Institute for Cell Science. For example, cells from a cancer biopsy might be assessed to see how things change as treatment is applied, or as a disease progresses. It can also help show how cells change within a growing organ or in the lab.
"This technique has huge potential ramifications for these and related fields," Horwitz said. "You can watch processes live as they are taking place -- it's almost like magic. This method allows us, in the most non-invasive way that we have so far, to obtain information about human cells that we were previously unable to get."
After training, a scientist can recognize some parts of the cell with brightfield microscopy. The cell’s edge, the nucleus, and some striking organelles like the Golgi apparatus might be identifiable. To train a computer to see more than that, the team used a convolutional neural network, an established machine learning method, to look for finer details. The team compared the results from the computer against fluorescent labels and found that most of twelve cellular structures were accurately identified. That team was surprised by the success of their technique.
"Going in, we had this idea that if our own eyes aren't able to see a certain structure, then the machine wouldn't be able to learn it," said study co-author Molly Maleckar, Ph.D., Director of Modeling at the Allen Institute for Cell Science. "Machines can see things we can't. They can learn things we can't. And they can do it much faster."
The same technique can be used with images generated by electron microscopy. Study co-author Forrest Collman, Ph.D., Assistant Investigator at the Allen Institute for Brain Science and an author on the study, is working with researchers that are mapping the links between mouse brain cells. This tool is enabling them to align images taken with different kinds of microscopes, normally a challenging and laborious task.
"Our progress in tackling this problem was accelerated by having our colleagues from the Allen Institute for Cell Science working with us on the solution," Collman said.
Roger Brent, Ph.D., a Member of the Basic Sciences Division at Fred Hutchinson Cancer Research Center, is applying this tool to a study he leads into improving the application of microscopy to yeast and mammalian cells. “Replacing fluorescence microscopes with less light intensive microscopes would enable researchers to accelerate their work, make better measurements of cell and tissue function, and save some money in the process," Brent explained. "By making these networks available, the Allen Institute is helping to democratize biological and medical research."
Sources: AAAS/Eurekalert! via Allen Institute, Nature Methods