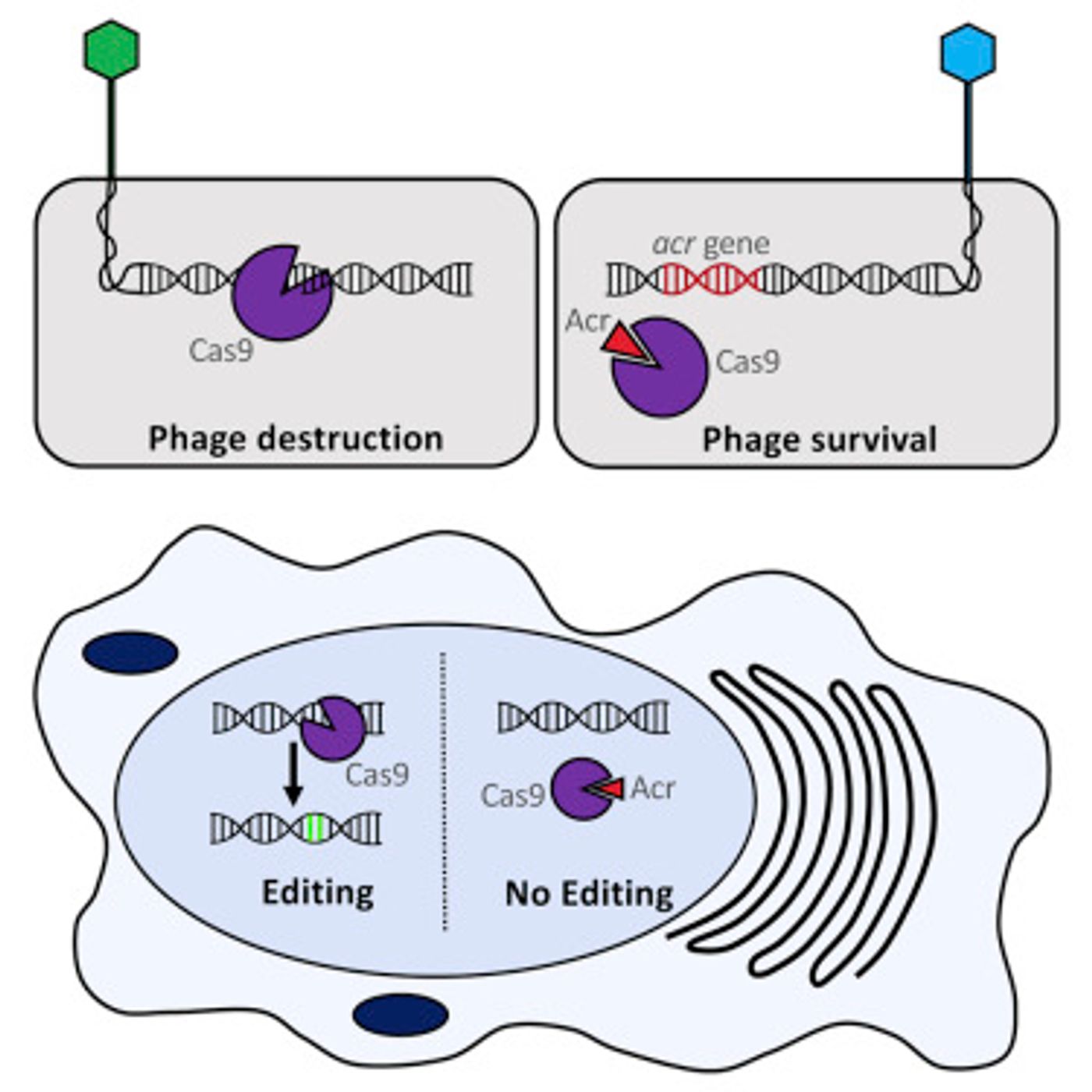
The CRISPR/Cas9 genome-editing tool set off a revolution in biomedicine. Natural features of bacterial immunity were utilized to make specific changes in the genome of a cell. Since its inception, many different variations and some improvements on the tool have been introduced. Now another discovery is set to add another dimension to CRISPR/Cas9 – scientists have found three naturally occurring proteins that can block cleavage of DNA by the Cas9 enzyme. The graphical abstract outlining the work is shown below where the Acr's are anti-CRISPRS that halt the enymatic cleavage.
One caveat of CRISPR/Cas9 is that it can make changes where they are not wanted – so called off-target effects, and scientists have found various ways to reduce them, but until now, there had not been a way to stop the action of the Cas9 nuclease. The
findings have been reported in Cell.
"CRISPR/Cas9 is a good thing because it introduces specific chromosome breaks that can be exploited to create genome edits, but because chromosome breakage can be hazardous, it is possible to have too much of a good thing, or to have it go on for too long," said one author of the work, Erik J. Sontheimer, a Professor in the RNA Therapeutics Institute at UMass Medical School. "There is a current shortage of reliable ways to turn off Cas9 once it has already been delivered to a cell. If you can trip an off-switch after the correct editing is done, then the problem is relieved. We report the first known natural inhibitors of Cas9 activity."
"Making CRISPR controllable allows you to have more layers of control on the system and to turn it on or off under certain conditions, such as where it works within a cell or at what point in time," said lead author Alan Davidson, a phage biologist and bacteriologist at the University of Toronto. "The three anti-CRISPR proteins we've isolated seem to bind to different parts of the Cas9, and there are surely more out there."
CRISPR/Cas complexes function naturally in bacteria to dice up the genomic material of invading viruses. Unsurprisingly, viruses have counteractive proteins that work to halt the actions of the CRISPR/Cas complex. Humans have hijacked the CRISPR/Cas system and made alterations to it; now the Cas9 enzyme that cuts DNA can be guided to a section of the genome that has been targeted for editing. That guide is not perfect though; it can make mistakes and go to the wrong places.
That major caveat is a hurdle in the laboratory, one that can be overcome, when performing experiments on cells or using animal models. But human subjects can’t afford to take major risks; we have to perfect the technique prior to using it as a therapeutic tool. This improvement is a big step in that direction.
"CRISPR-Cas9 in ancillary cells, tissues, or organs is at best useless and at worst a safety risk," said Sontheimer. "But if you could build an off-switch that keeps Cas9 inactive everywhere except the intended target tissue, then the tissue specificity will be improved."
"Knowing we have a safety valve will allow people to develop many more uses for CRISPR," commented co-author Karen Maxwell, an Assistant Professor in Biochemistry at the University of Toronto. "Things that may have been too risky previously might be possible now."
There is much more to come from this research. "There are many different forms of Cas9 that are produced by different bacteria, and the different Cas9s can each have different useful properties in genome editing," Sontheimer said. "So there are already several Cas9s in the toolbox, with many more to come. We've now proven that such inhibitors are out there in the natural world and we've provided one possible strategy to find them."
Davidson is interested in delving into the bacterial side of the CRISPR and viral anti-CRISPR molecules interact. "We didn't set out to find anti-CRISPRs, we were just trying to understand how phages incorporate themselves into bacterial genomes and stumbled onto something that I think will be important for biotechnology," Davidson says. "We were being observant and following a path that we didn't know where it could lead, and it's just been a very fun and exciting story."
The video above from the National Science Foundation discusses how research on bacteria led to discoveries about CRISPR/Cas9.
Sources:
AAAS/Eurekalert! via
UMass Medical, via
Cell Press,
Cell