Shedding Light on a Genomic Mystery
There are untold numbers of bacteria in our world, and we play host to trillions of microbes. It was once thought that bacterial cells outnumbered human cells in our body, and while that has been revised to a ratio of about one to one, it still means we carry far more bacterial genes than human genes. However, we know very little about what those bacterial genes do. Researchers want to shed light on this so-called ‘genomic dark matter.’ An international team of researchers has reported a new way to predict bacterial gene function in the journal Microbiome.
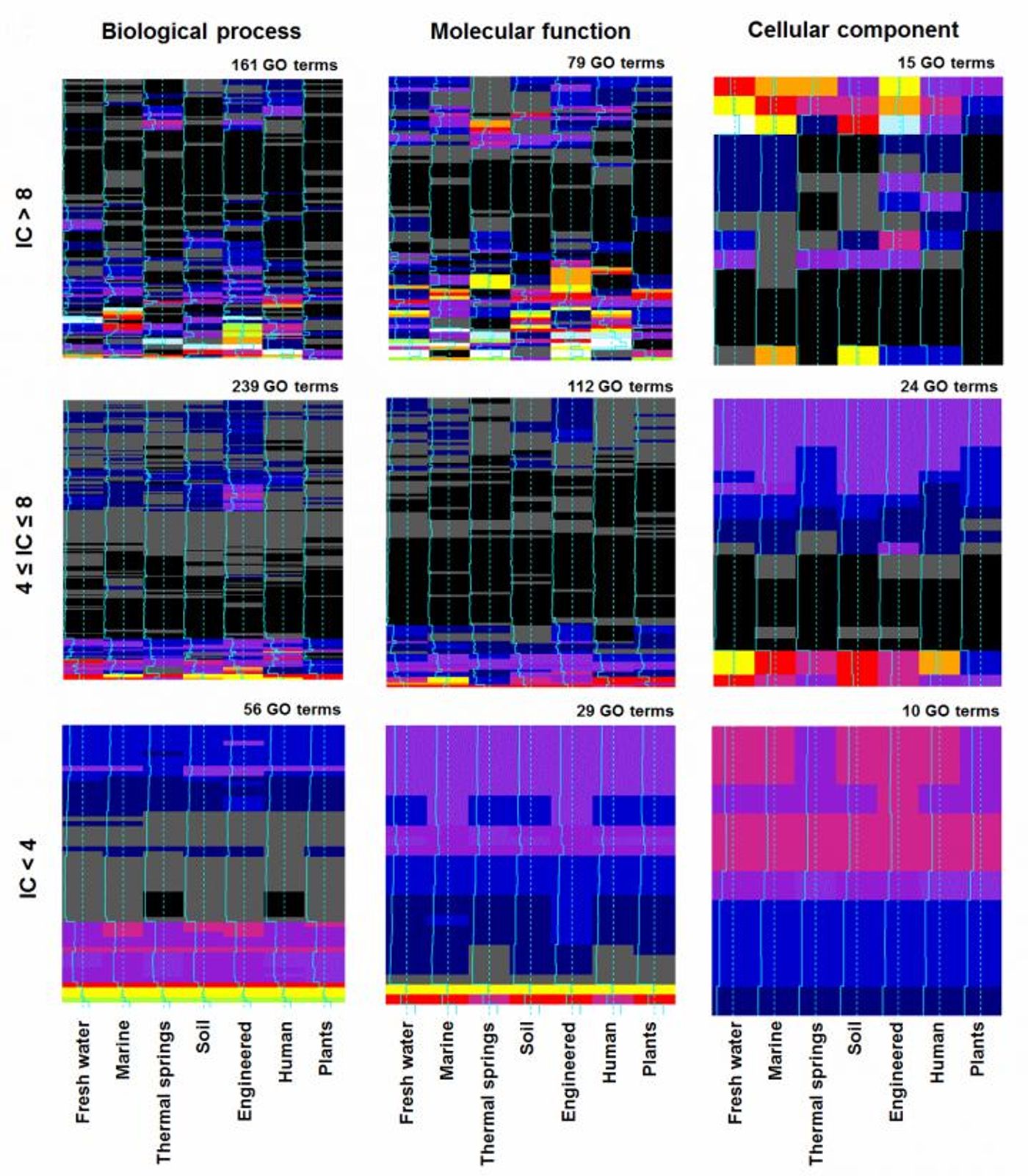
The team created a method to use computation to assess a tremendous amount genetic material from bacteria - thousands of metagenomes - at the same time. This technique can be used to analyze bacterial communities hosted by humans, like the skin or gut microbiomes, or other kinds of metagenomes, like those of the ocean or soil. The aim is to find an evolutionary signal to predict the role of a gene. The tool uses machine learning, which generates decision trees, linking genes to function, and suggesting what their role is in a microbial cell.
"This makes the algorithm very good at not getting confused by the noise in the metagenomic data, meaning that it is accurate and can confidently propose a biological role for a large number of genes with unknown functions. Intriguingly, it also proposes many additional functions for genes that already have some known role," noted study leader Fran Supek, computational biologist and leader of the Genome Data Science lab at IRB Barcelona.
This study has opened up new avenues to understand microbial gene function and has given researchers a new way to look at gene function in the microbiome. "In other words, metagenomes allow scientists to see what ordinary genomes don't," explained Supek.
This work has suggested that environments help predict gene function. Metagenomes originating in the ocean, for example, can help reveal genes that bacteria use in photosynthesis. The researchers note that gut bacteria would not provide this kind of information - photosynthesis would be a useless function in the human GI tract. The gut microbiome is useful for predicting genes that are involved in metabolic function, like amino acid biosynthesis or the breakdown of alcohol. Microbes in the environment, in turn, are unlikely to be using those kinds of genes.
"Computational methods like this one are shedding light on the "dark matter" within microbial genomes - the enormous number of genes in bacteria and in archaea whose functions are a mystery," said Supek.
Although they have made thousands of computational predictions, these findings will still have to be validated. That validation will help show how bacteria exert their influence on the ecosystems they inhabit, including the human body.
Supek is featured in the video above discussing his research.
Sources: AAAS/Eurekalert! Via Institute for Research in Biomedicine, Biochemical Journal, Microbiome