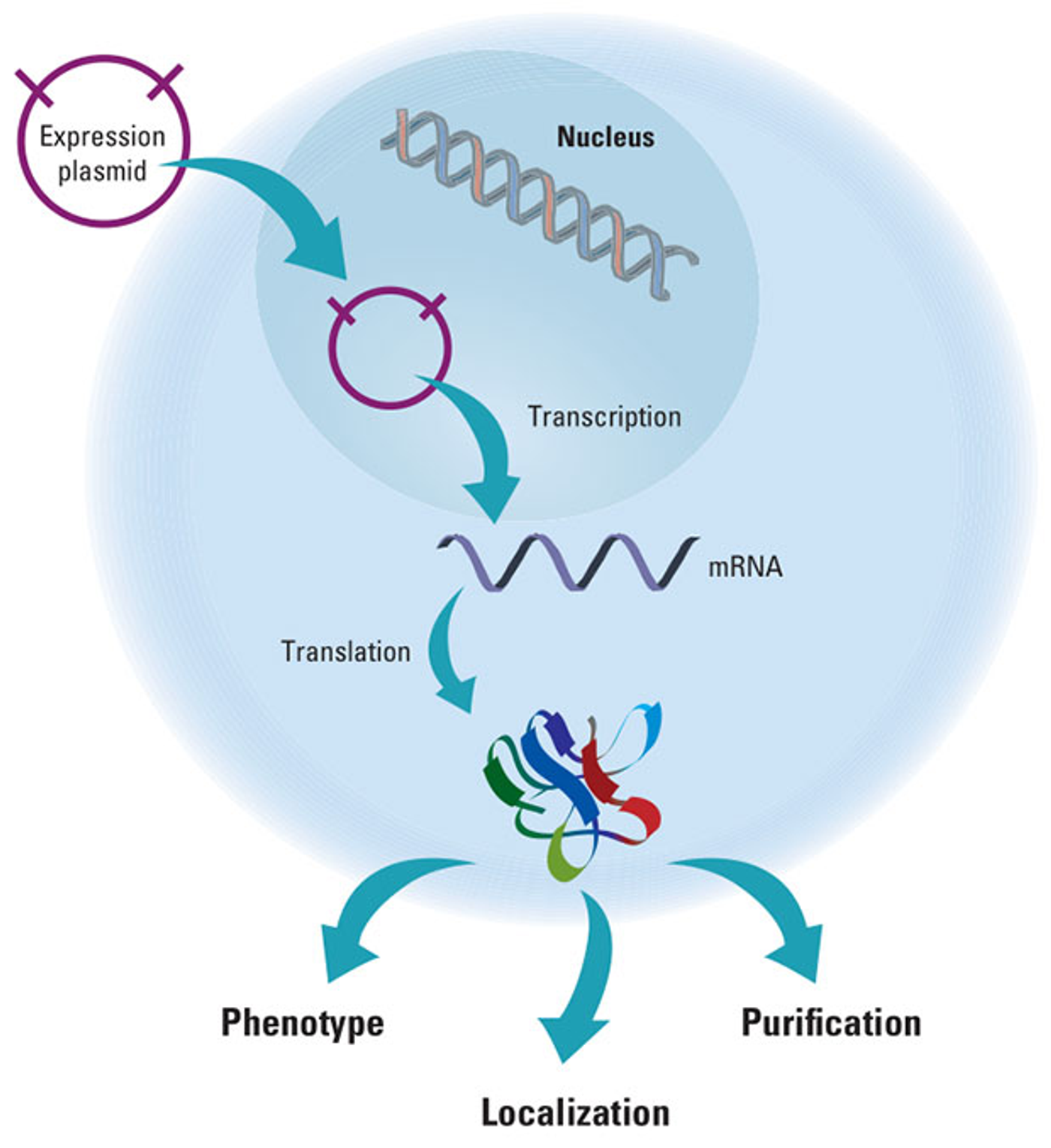
Gene expression is more than just being on or off. It is more of a spectrum of expression, scientists are discovering. In a new study published in
Nature Genetics, Garth Ilsley, PhD, from the Okinawa Institute of Science and Technology Graduate University (OIST), used a mathematical model to predict how and where genes are expressed using artificial transcription factors in fruit flies.

Transcription factors bind to specific sections of DNA called enhancers, either inhibiting or activating expression of the particular gene for which the DNA codes. Ilsley likens the interaction between transcription factors and their enhancers to the tuning of a radio.
“Some transcription factors turn the volume up, while others turn it down.”
Ilsley’s model measures how transcription factors bind to enhancers, an important quality for his studies since each transcription factor binds in a unique way. In addition to predicting the final volume of expression, the model also predicted where each gene would be expressed. Focusing on fruit fly segmentation, Ilsley and his team used artificial transcription factors with different binding strengths to test the efficacy of the model, and they were successful.
He found that the expression predicted during his experiment was both controllable and reproducible, qualities important for applications in “stem cell reprogramming and regenerative medicine.”
Ilsley’s study also has an evolutionary significance. While using artificial transcriptions factors, he noticed flexibility in the enhancers’ ability to accept different kinds of transcription factors in different ways.
"You can bring in foreign transcription factors and the enhancers still work,” Ilsley said. “The enhancers we looked at are not brittle at all.”
This ability of enhancers means that the same enhancer can work differently in different situations over time, he continued. Just like a radio continuously has to be tuned to be in sync with local broadcasts, transcription factors and enhancers change in the way they bind and influence each other depending on their local cellular environment.
Source:
Okinawa Institute of Science and Technology Graduate University