Some mutations in the DNA are harmless, and a person could go their entire life with the mutation and never know. Other mutations cause a myriad of health problems: cancer, diabetes, heart disease, neurodegenerative disorders, autoimmune and inflammatory disease. Early detection and potentially even prevention of many of these diseases might now be possible with a new electronic and genetic system for identifying disease-causing mutations.
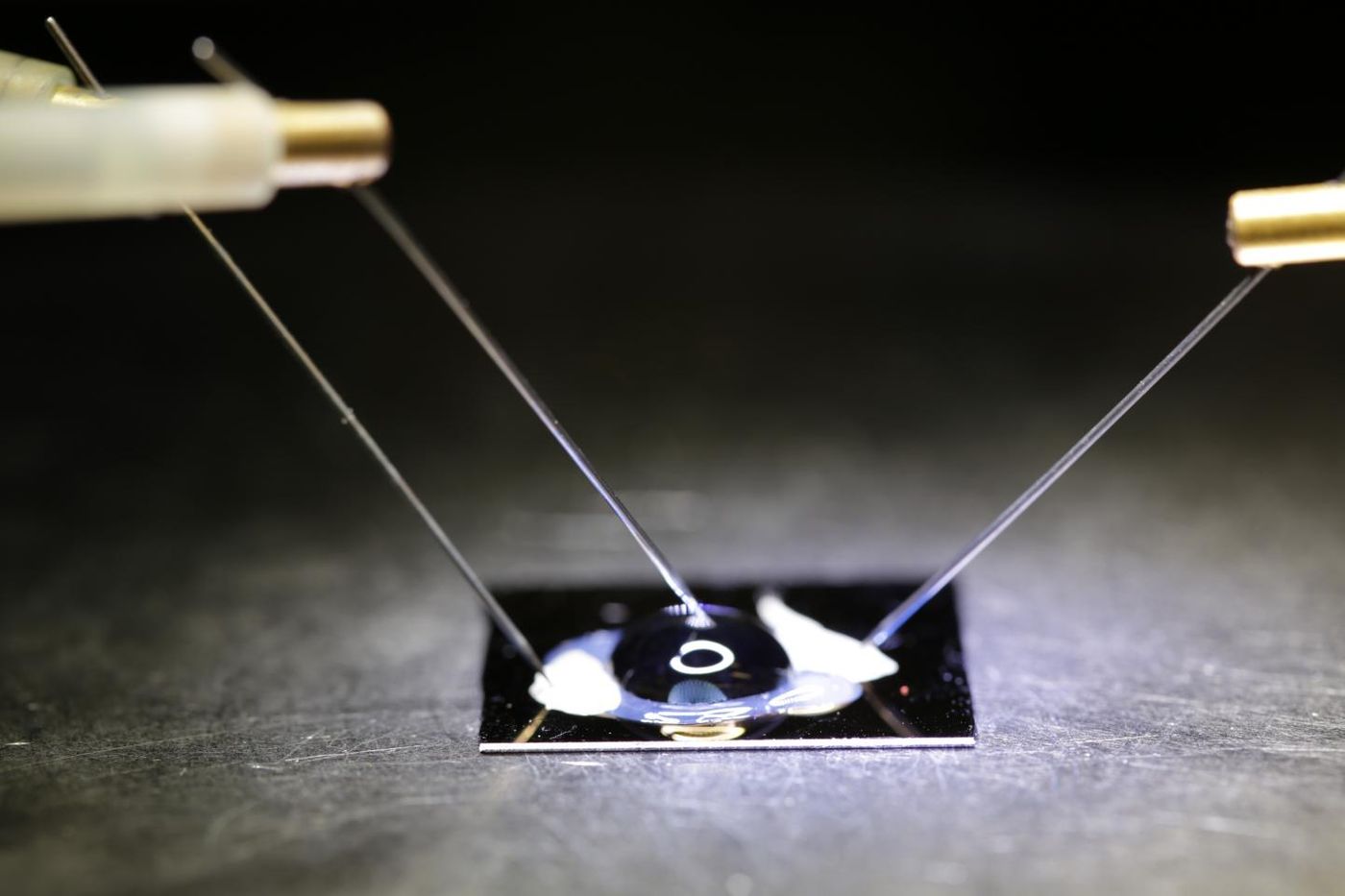
Scientists know a lot of the specific mutations that cause disease, and they continuously look for different ways to apply their knowledge to more than just making diagnoses. A newly developed technology consisting of a double-stranded DNA probe attached to a graphene field effect transistor could be the new way to sense specific mutations known to cause disease. The biosensor chip is currently being fine-tuned by bioengineers at the University of California San Diego.
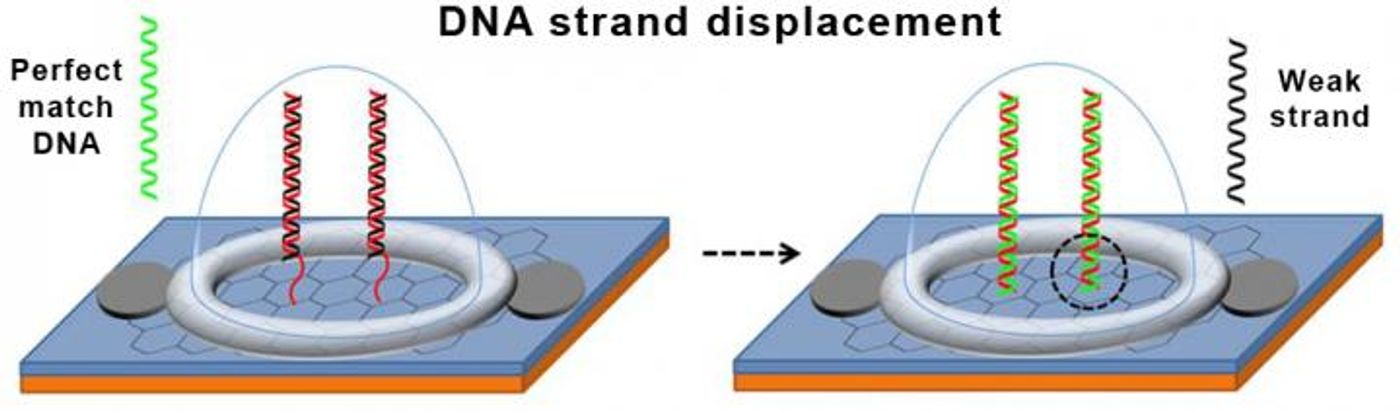
The novel design of the DNA probe provides better than ever accuracy. One strand of the double helix is attached to the graphene field effect transistor. This “normal” strand is complementary to a specific strand of DNA containing a single nucleotide polymorphism (SNP), a common genetic mutation. The other strand of the double helix has four defective nucleotide bases – guanines that were replaced with inosines so the strand would bind to the normal strand, but not as strongly as a truly complementary strand would bind.
In theory, once the biosensor chip is inserted into the body, if it were to come into contact with the truly complementary strand to the normal strand of the DNA probe the defective strand would be replaced with the complementary strain from the body. At this point, the sensor is designed to produce an electrical signal that the researchers intend to be transmitted wirelessly to a mobile device.
“This is the first example of combining dynamic DNA nanotechnology with high resolution electronic sensing, said Michael Hwang, a PhD student at the University of California San Diego and a co-first author of the study. “The result is a technology that could potentially be used with your wireless electronic devices to detect SNPs.”

The future of this technology is as vast as scientists’ understanding of the genetic mutations that cause certain diseases. If they know the SNP that causes a certain autoimmune disease, they can create the complementary strand and attach it to a biosensor chip to be sent into a patient suspected to have the disease or has a family history of the disease. The researchers from the study, published in
Proceedings of the National Academy of Sciences, anticipate the biosensor chip to be useful for “blood-based tests for early cancer screening, monitoring disease biomarkers, and real-time detection of viral and microbial sequences.”
Compared to current and past SNP-detection technology, the new biosensor chip is expected to be faster, more accurate, and less expensive. Simply because the design incorporates double-stranded DNA as opposed to single-stranded DNA, this novel technology ensures that only the perfect match will cause a test to read positive because of the need for the complementary strand found in the body to not only bind to the DNA probe but also to displace the defective complementary strand on the probe.
The DNA probe can also detect SNPs as long as 47 nucleotides, a length greater than any previous SNP-detecting technologies were capable of detecting. Researchers say that this biosensor chip is in the “proof-of-concept” stage still, but once it has been tested in clinical studies, scientists can expect the chip to jump-start a “new generation of diagnostic methods.”
Sources:
University of California San Diego