Bacteriophages are viruses that infect bacteria. Have you ever wondered how these guys find their bacterial “prey”? Investigators at San Diego State University demonstrated that “subdiffusive motion” helps phages efficiently find bacteria.
The San Diego State University study investigated the
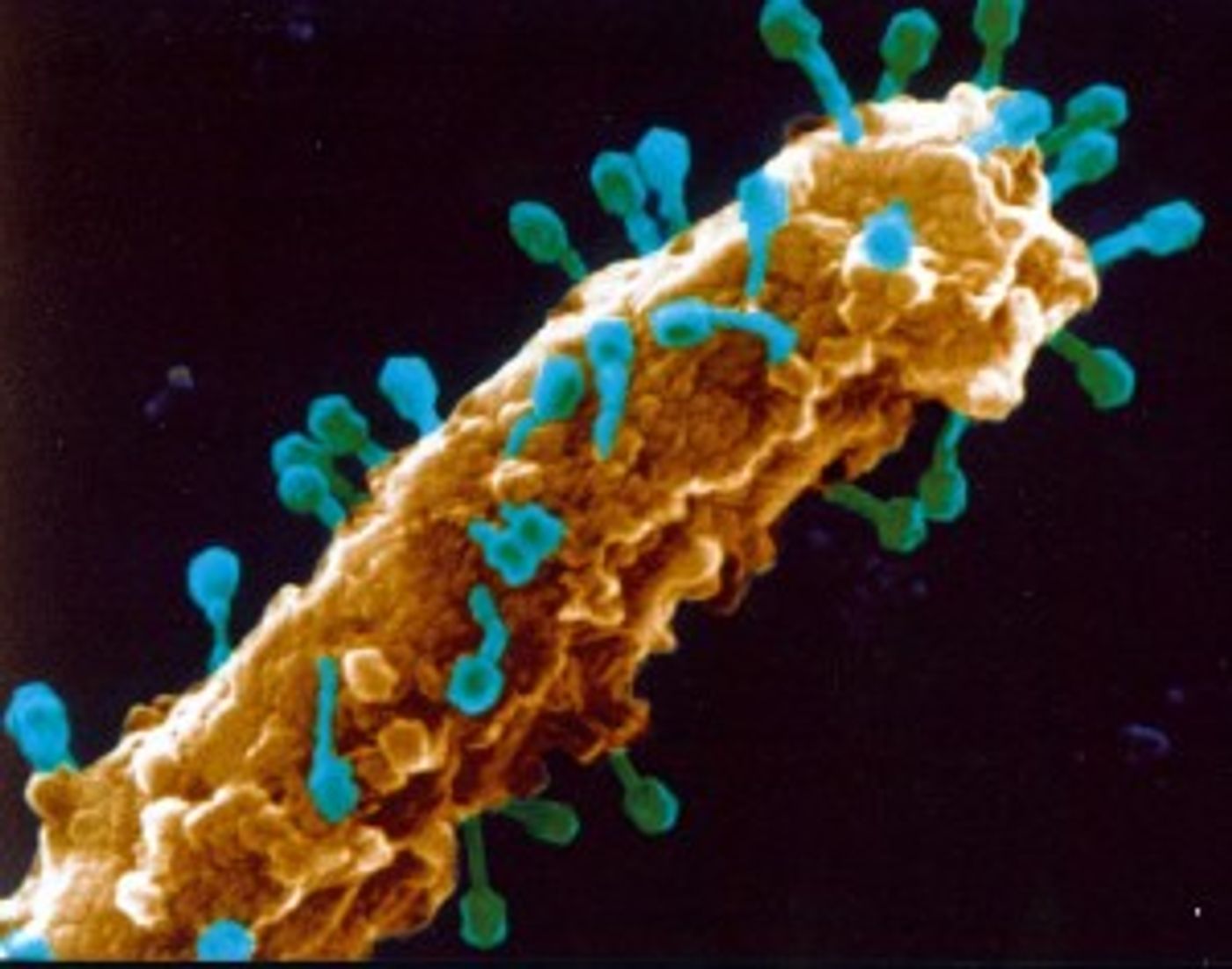
T4 bacteriophage, a lytic phage that infects
E. coli. T4 is one of the more recognizable phages, somewhat resembling a lunar lander. The T4 genome is made up of double-stranded DNA and encodes 289 proteins. The DNA is housed in an icosahedral head, or “capsid”, and long tail fibers recognize receptors on the bacterial cell surface. The tail also serves as a hypodermic needle through which phage DNA is inserted into the host cell.
Until now, the so-called “BAM” model (bacteriophage adherence to mucus) has been used to describe how phages move through mucus to encounter bacteria. In the BAM model, phages randomly diffuse through mucus by Brownian motion, but lead investigator Jeremy Barr wondered if the process was actually more complex.
First, Barr and colleagues used a biochip that mimicked the mucus layer on lung epithelial cells to characterize phage “hunting strategies”. They added
E. coli to the mucus layer along with wild type T4 phage, or mutant T4 that could not adhere to mucus. They found that the bacterial load decreased by nearly 4,000 fold when phages were able to adhere to mucus, but mutant phages were not able to kill bacteria.
Next, the team used microscopy to watch phages diffuse through the mucus. They found that wild type phages (that were able to adhere to mucus) moved by “subdiffusive motion”. With this type of movement, phages move a short distance, pause by adhering to mucus, then diffuse on. It appears that this mode of travel is most effective when bacterial concentrations are low, or diversity is high (Brownian motion is sufficient when bacteria are abundant). According to Barr, “when bacterial diversity is high, it becomes difficult for a phage to find its specific host … under these conditions it’s more advantageous to move subdiffusively, to keep yourself in a location longer and seek out your prey more slowly”.
This is exciting work, because these findings could help researchers engineer phages that are exceptionally good at killing bacteria. Such phages could then be used in phage therapy to treat bacterial infections.
Sources:
Phys.org,
PNAS, Wikipedia