Small Differences Enable Some Microbes To Persist in the Gut
The world is full of microbes that have colonized nearly every environment including the human body. The vast majority of these microorganisms are harmless, and some are even important to maintaining our good health. But some are pathogenic. Certain strains of bacteria can cause serious illness, some of which may be temporary while others can cause chronic diseases like ulcers, or cancer, or even death. But it appears that seemingly minor differences can make one strain of bacteria dangerous while another is harmless, even the same type of microbe can have pathogenic and non-pathogenic strains.
Helicobacter pylori is a bacteria that might live in the stomachs of people without causing any problems at all, but in others, it can invade epithelial cells; it may lead to chronic inflammation, ulcers, and potentially, cancer. Scientists have now investigated the molecular characteristics that make some strains of H. pylori adapt to living in the human stomach, and causing disease, for a long time.
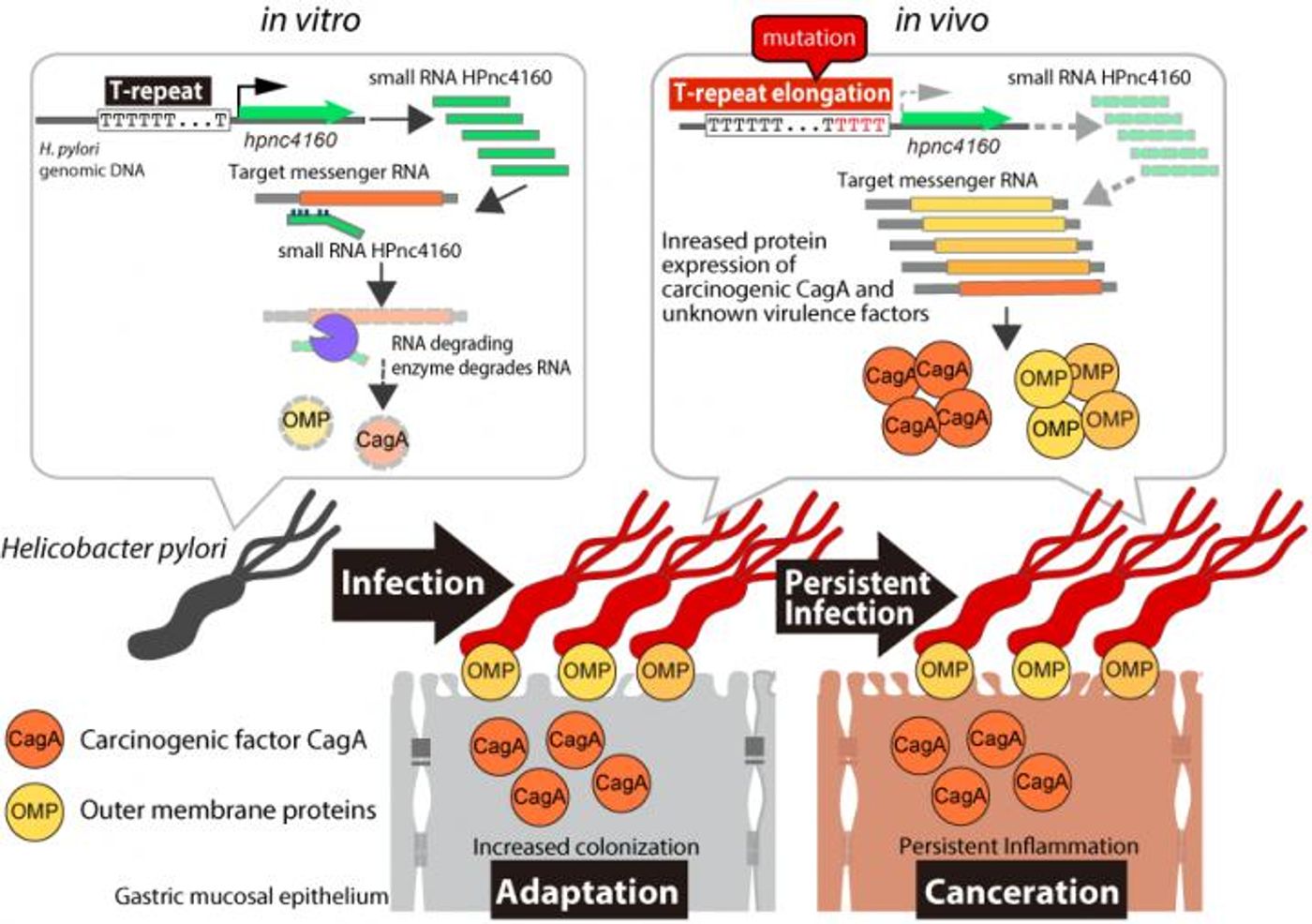
Reporting in Nature Communications, the researchers identified a small RNA molecule called HPnc4160, which has an important role in how H. pylori behave in the stomach and can cause disease.
Previous research has shown that different strains of H. pylori produce HPnc4160, and since the microbe retains the gene, that suggests its function is important. However, different strains of H. pylori produce HPnc4160 at varying levels. An analysis of the genetic sequence of these microbes indicated that one part of the T-repeat region in the H. pylori genome has varying lengths in different strains. This region was linked to a change in HPnc4160 expression levels, and also seems to make some strains better at colonizing the human stomach than others.
"Slight differences in the genetic sequence of H. pylori can sometimes give certain strains advantages over others that allow it to grow better," explained the lead author of the study Ryo Kinoshita-Daitoku. "We were interested in the genetic and molecular reasons behind why particular strains are more pathogenic and can live in the stomach for decades, leading to cancer development."
In this study, gerbil and mouse models were used to study H. pylori infections. After eight weeks of infection, the genomes of the microbes were sequenced. The researchers found that strains of H. Pylori with higher levels of T-repeats had reduced HPnc4160 expression.
"We found that the H. pylori strains with low HPnc4160 levels were more infectious," said the senior study author Hitomi Mimuro. "When this RNA was completely absent, the amounts of several bacterial pathogenic factors significantly increased and the strain showed stronger colonization within the rodent stomach."
Another pathogenic influence was a bacterial protein called CagA, which is known to promote the development of cancer.
"Interestingly, we observed longer T-repeat regions, lower levels of HPnc4160, and higher levels of CagA in gastric cancer patients compared with those not suffering from cancer," added Kinoshita-Daitoku.
This work could help researchers create better treatments for H. pylori infections.
Sources: AAAS/Eurekalert! via Osaka University, Nature Communications