A sneaky way to antibiotic resistance
If you’re a bacterium, you don’t need to be resistant to antibiotics to survive them. You just have to make friends with bacteria that are. For susceptible cells that are in the right place at the right time, antibiotics don’t have to mean death.
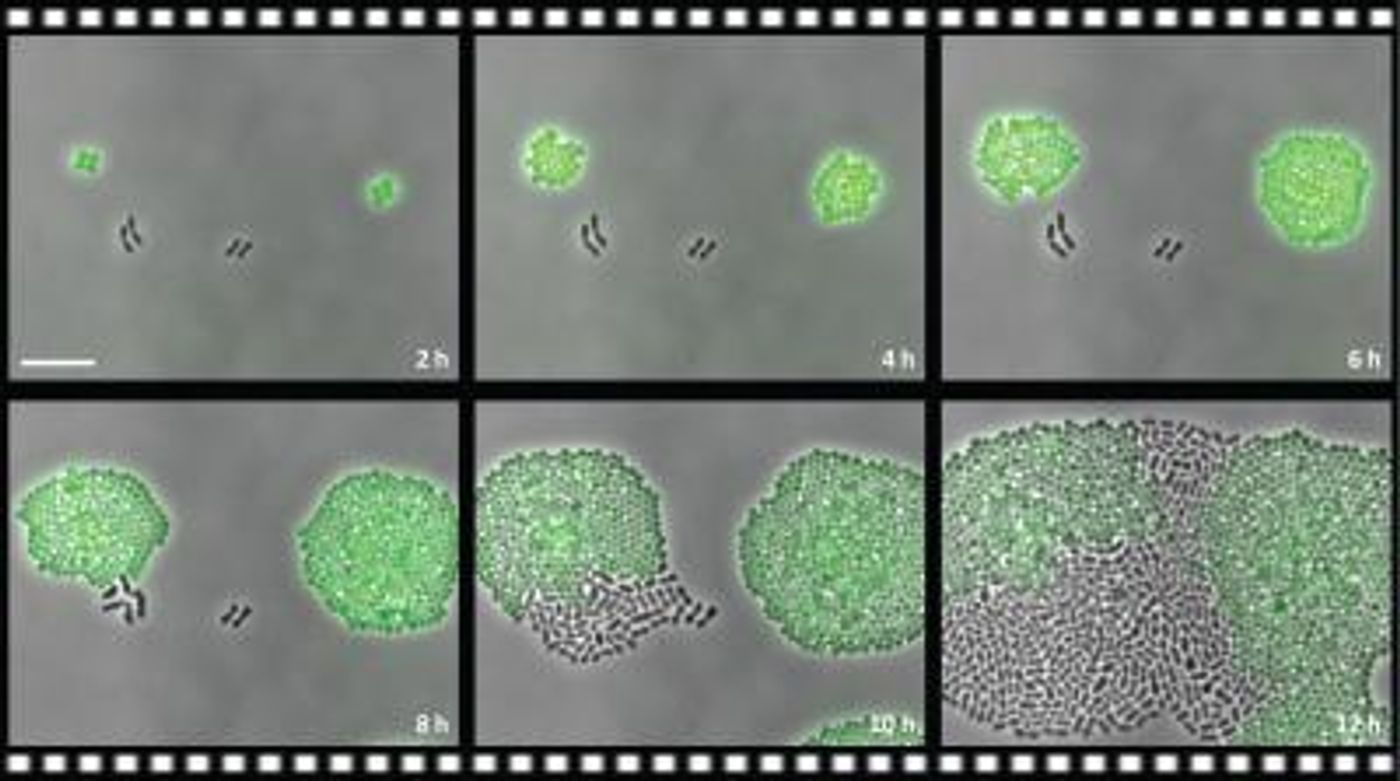
Researchers from the University of Groningen showed that chloramphenicol-sensitive Streptococcus pneumoniae can grow if they are placed next to chloramphenicol-resistant Staphylococcus aureus - the resistant bacteria use the enzyme chloramphenicol acetyltransferase to inactivate the antibiotic, making the area habitable for the susceptible Streptococci.
This differs from the way in which penicillin can be broken down by secreted beta-lactamase enzymes. According to study author Robin Sorg, “cells with resistance to penicillin can secrete beta-lactamase enzymes which break down the antibiotic. But in our case, the antibiotic is deactivated inside the resistant cells.”
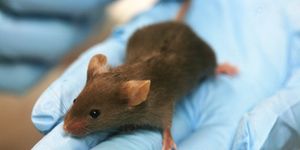
The researchers wanted to know if the chloramphenicol-resistant cells could rescue the growth of sensitive cells in the presence of chloramphenicol (Cm) in an animal model. They used a lung infection model that allowed them to quantify bacteria present in the lungs of mice. When the mice were infected with Cm-sensitive (CmS) bacteria alone or with both CmS and Cm-resistant (CmR) bacteria, there were no significant decreases in the number of viable bacteria recovered from the lungs after 24 h. When the mice were infected with CmS bacteria and treated with Cm, the antibiotic significantly decreased the number of CmS bacteria recovered from the lungs compared to an untreated control, as expected.
However, when the mice were infected with both CmS and CmR bacteria and treated with Cm, there was no significant decrease in the number of viable bacteria - consistent with their in vitro findings. In fact, 6 of the 14 animals ended up with more CmS cells in their lungs than CmR!
Importantly, these CmS cells didn’t survive because they acquired the chloramphenicol-resistance gene from the resistant bacteria - they were not able to grow in Cm by themselves after being recovered from the animals.
These findings point out how the presence of multiple types of microbes can complicate antibiotic therapy. These types of studies could also help explain how antibiotic resistance develops, and Sorg thinks her work could help doctors better treat patients with antibiotics. “We know that antibiotic usage results in selection for resistance. However, we do not fully understand the processes, nor why antibiotic resistance can develop so fast. Single cell studies like ours help to fill in some of these details. We know that we should use these drugs with discretion, but we may need to be even more careful than we thought,” says Sorg.
Sources: PLOS Biology, EurekAlert