Researchers in Japan have created a protein that fluoresces and assembles into large crystals when placed inside a living cell. The team, from the RIKEN Brain Center, showed that these cells then actively targeted the crystals for degradation. Because they were so large and pure, unlike anything placed in a cell before, it was possible to fully analyze the protein's structure within an intact cell. The researchers were able to observe the crystallization process and the cell's responses in real-time, potentially transforming the production and study of protein crystals.

Dr. Atsushi Miyawaki led the team from RIKEN and their research was published online in the journal Molecular Cell. They were success in genetically engineering a fluorescent protein that can rapidly self-assemble into well-formed single crystals-highly ordered structures composed of individual units arranged in a repeating lattice-in live mammalian cells, based on a sample of a fluorescent coral brought back from Okinawa.
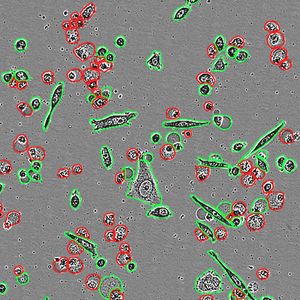
In addition the team was able to isolate the gene encoding in the green fluorescent protein, called KikG, and manipulated its structure so that the color changed from green to red when struck by a particular wavelength of light. They also observed the emergence of micron sized fluorescent crystals in the cells used to make the proteins. This was not an expected result by the team. They named these crystals Xpa.
Prior to the crystallization, the green-colored Xpa proteins are distributed evenly throughout the cell. A video of the process shows a large fluorescent crystal appearing suddenly within one frame of the video but it actually happened much faster. The camera was set at ten-minute intervals, but the process of crystallization happened within a minute. Once crystallization begins, individual Xpa proteins are rapidly recruited into the growing crystal until the cell's cytoplasm becomes clear.
Miyawaki state, "This was quite a fortuitous discovery, because the ability to crystallize proteins makes it possible to determine their atomic structure." Prior to the RIKEN team's discovery, creating a crystal big enough for analysis required weeks or months, and sometimes, despite best practices, no crystal would form. In contrast, the crystals grown by the RIKEN team appeared in minutes and were so pure that they could be examined directly, without being removed from the cell, at RIKEN's SPring-8 facility. This is the first time such a feat was accomplished in animal cells, opening the door to analysis of protein structure directly inside cells.
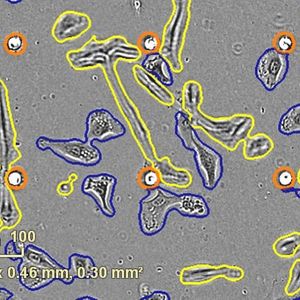
A second significant finding was that after the crystals formed, the cells attempted to destroy them through a process known as autophagy. "There may be evolutionary pressure to discourage crystal formation in mammalian cells," said Miyawaki. The group discovered that the crystals were rapidly covered by a membrane structure called the lysosome, normally an acidic, digestive compartment of the cell. Usually, proteins are efficiently broken down by the lysosome, however, because Xpa originally came from coral, the crystals could not be digested and remained in the cell and the attempt at autophagy was not successful.
Miyawaki believes there may be undiscovered protein crystals inside cells that can be either beneficial or harmful to the cell, but have been overlooked because they are not fluorescent. "There may be similarities between Xpa protein crystallization and the formation of pathogenic protein aggregates in diseases like Alzheimer's," said Dr. Miyawaki. "We hope to develop fluorescence detection systems for crystal growth to screen for molecules that affect the cell's response to pathogenic protein aggregates, with the hope that this will lead to medical advances in the future."