Predicting the Impact of Gene Splicing Errors
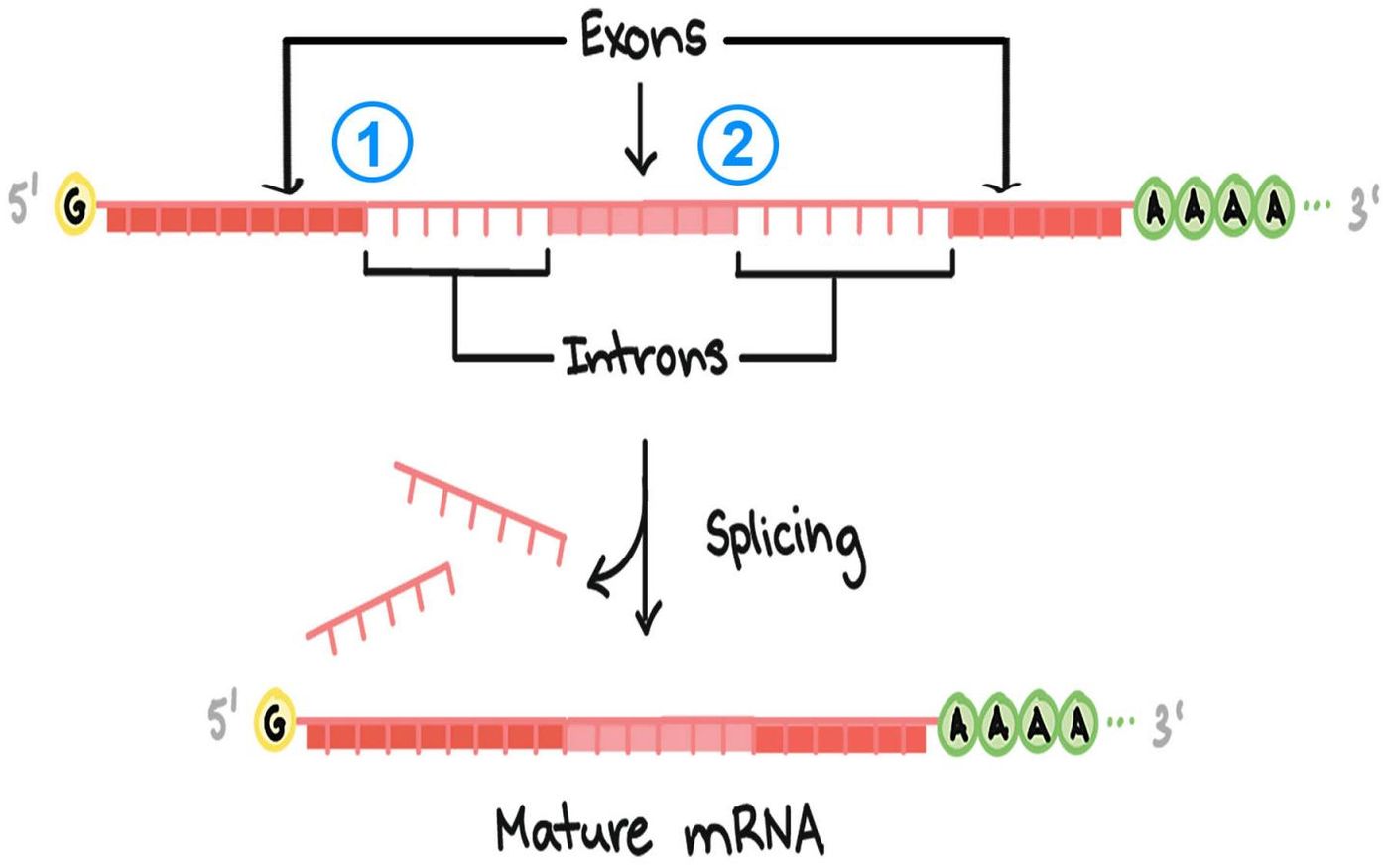
Our cells have to generate a massive number of proteins, and there is a carefully orchestrated procedure for doing so. It starts when active genes are transcribed into RNA, and while that RNA eventually gets translated into proteins, the RNA undergoes many edits before that happens. One of those edits is splicing, in which parts of the RNA are removed (the process is described in the following video). Researchers at Cold Spring Harbor Laboratory (CCHL) are turning their attention toward that splicing; they want to know more about how it may play a role in disease when it is not functioning correctly.
Genes have different sections; one of those codes for protein - the exons. Those areas are interspersed with non-coding sequences, called introns. Cell have to remove, or splice out, the introns. If that splicing happens incorrectly, it might have a devastating impact on the resulting proteins. It’s been estimated that 14 percent of disease-causing mutations that trace back to single base pair changes in RNA are related to intron sequences.
At Cold Spring Harbor Laboratory (CSHL), scientists led by Professor Adrian Krainer and Assistant Professor Justin Kinney are studying how RNA splicing is controlled in cells. They want to connect genetic mutations with changes in that process and use that information to predict disease risk in individuals. New work on that front has been reported in Molecular Cell.
There are a variety of splice sites throughout the genome, and splicing errors can have a wide array of effects. When a splice site sits near the start of an intron, called the five prime (5’) end, Krainer noted that "we know that at the first and second [DNA-letter] position, mutations have a very strong impact. Mutations elsewhere in the intron can have dramatic effects or no effect, or something in between."
Predicting how a mutation at a splice site will affect patients has been challenging. For the BRCA1 or BRCA2 genes, which are strongly linked to breast cancer, any gene mutation raises the breast cancer risk, but, not every mutation in those genes turns out to be harmful.
For this work, Krainer lab postdoc first author Mandy Wong and colleagues engineered splice sites with every combination of sequences at the 5’ end of the gene to assess the removal of associated introns. They did this for three genes linked to disease, the breast cancer gene BRCA2 as well as IKBKAP and SMN1, which have been linked to neurodegenerative diseases.
The team ultimately tested more than 32,000 5' splice sites in one intron, for every gene. Certain sequences correlated with changes in splicing efficiency, some increasing and some decreasing it. Other parts of the gene also had an impact. The researchers suggest that it is possible to predict the effect of 5’ splice site mutations, but only in the context of the gene.
While this data enhances what we know about predicting the effects of splice site mutations, more work will be needed.
Sources: AAAS/Eurekalert! Via Cold Spring Harbor Laboratory, Molecular Cell