Proteins are the building blocks of the cell through which vital cellular functions are carried out. Remarkably, there are more than 100,000 proteins in our bodies produced from only 20 amino acid building blocks. In humans these functions include skeletal structure support, control of the senses, muscle movement, digestion of food, and defense against pathogens and infection. In order for each protein to properly perform its function, the protein must be correctly folded into a complex 3D structure.
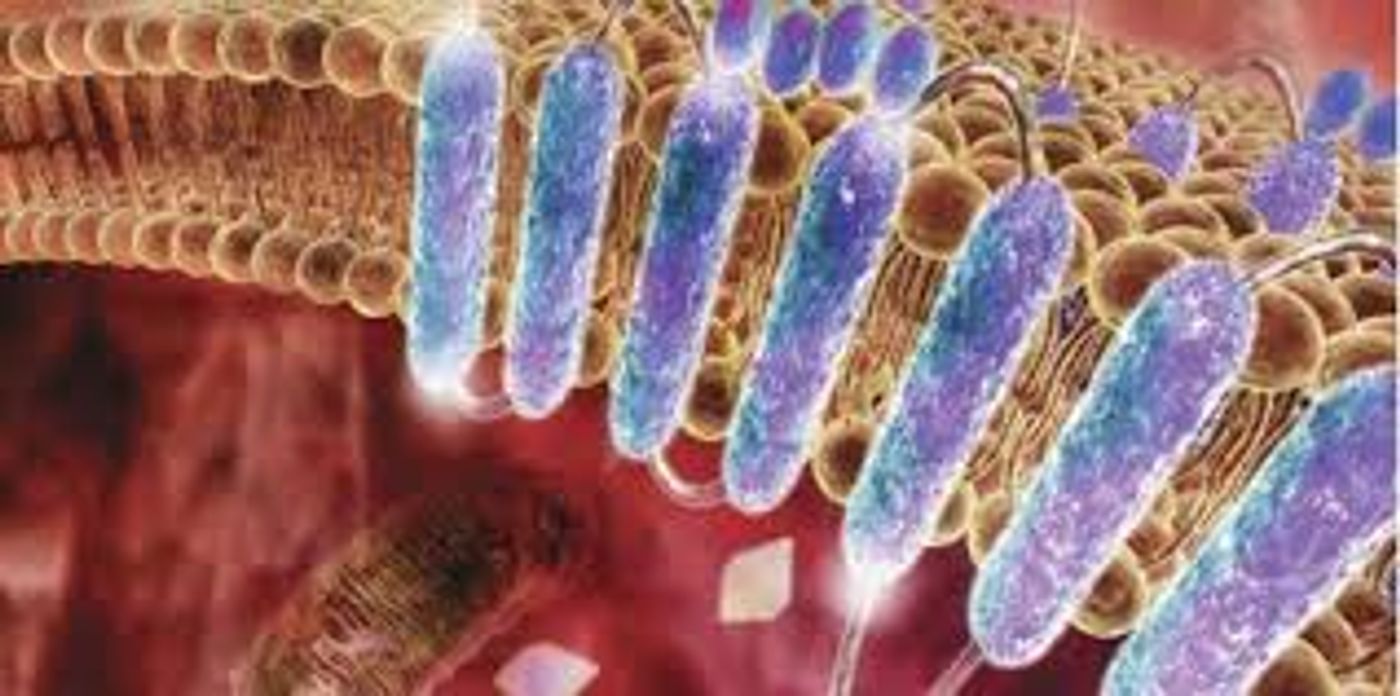
Within the cell membrane, proteins are designed to fold and function within the lipid bilayer. The method by which proteins in the cell membrane are folded can be broken down into two major stages: (1) Initial insertion of transmembrane helices and (2) completion of protein folding into its final native structure. Scientists have hypothesized, based on this concept, that membrane protein folding would be performed ideally in the membrane, however; performing a study to confirm this would require altering the energy landscape so that it would favor the unfolded state of the protein. While there are techniques available to force unfolding of membrane bilayer proteins, such as atomic force microscopy (AFM), this technique applies force parallel to the membrane normal so that proteins are physically pulled out of the membrane. To study folding within the cell membrane, a technique to apply force along the membrane plane would be necessary.
Researchers from the Department of Physics at the Korea Advanced Institute of Science and Technology (KAIST) and the Department of Chemistry and Biochemistry at UCLA have developed a new method that allows manipulation of folding membrane proteins using a magnetic tweezer. To do this, scientists first attach long DNA handles to the ends of a protein. One DNA handle is attached to a glass surface and the other is attached to a magnetic bead. Using the magnet, they are able to “pull” on the protein itself, allowing it to fold and unfold. They are able to visualize this using 3D tracking of the magnetic bead.
Using this novel technique, the research team was able to quantitatively map the folding energy landscape, kinetics of folding, and folding intermediates of a protein located within the cell membrane for the first time. The folding landscape of the membrane protein used in this study included high cooperativity, low thermodynamic stability, a high kinetic barrier and a transition state that was structurally more similar to the folded state than the unfolded state. The authors of the study report that they are unsure whether these landscapes can be applied across all membrane proteins. This new technique will allow scientists to study folding characteristics of other membrane proteins in the future.
Sources:
Nature Chemical Biology;
Science Daily;
Nature Horizon Symposia