Antibiotic resistance has become one of public health’s largest issues in recent years. A recent study from the UK suggests that antibiotic resistance could become a leading cause of death by 2050, which would surpass cancer. New evidence from a multi-disciplinary group led by the University of Oklahoma with the Department of Energy's Oak Ridge National Laboratory, the University of Tennessee and Saint Louis University, might just be able to halt this trend.
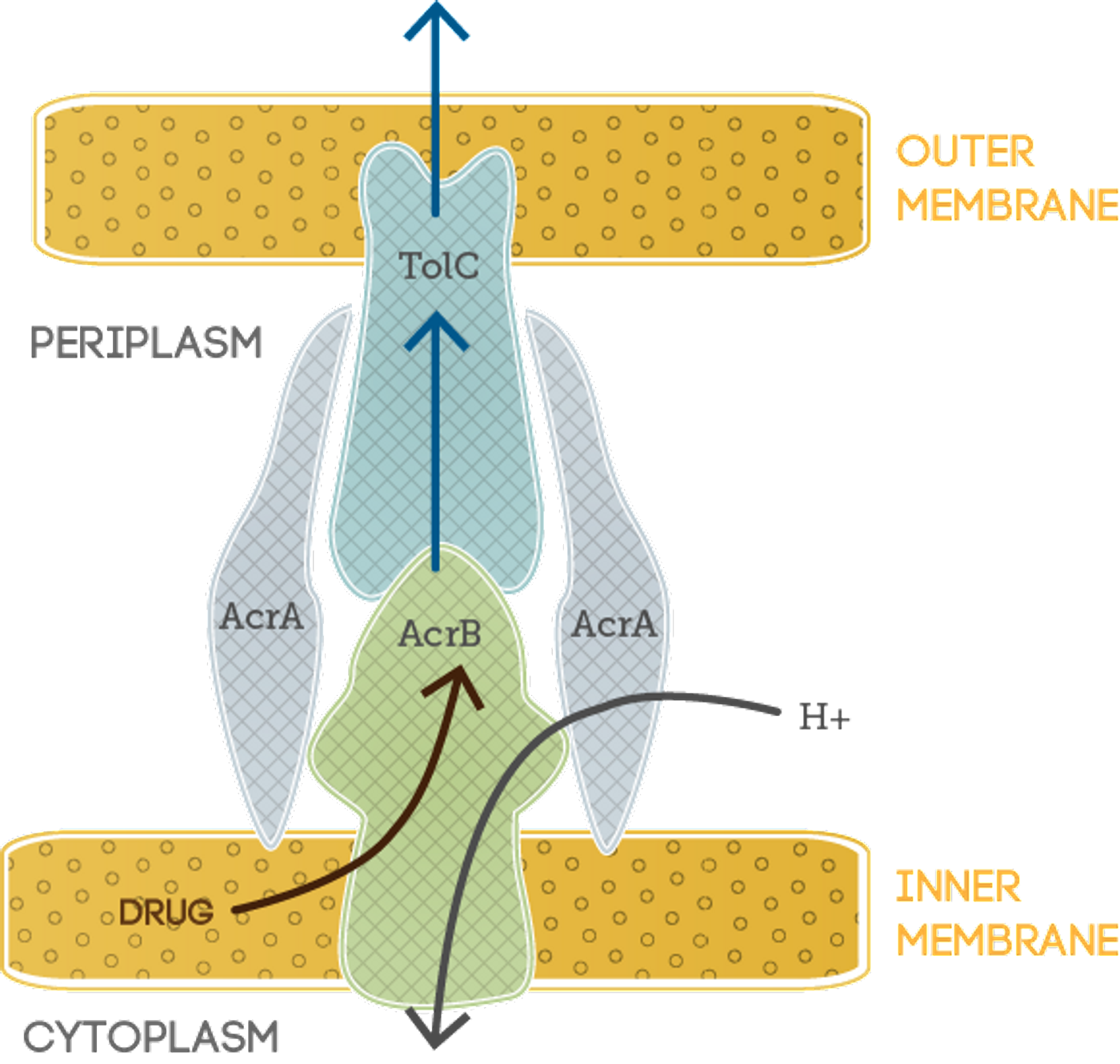
The team is contributing to an alternate approach to looking at drug resistance. Instead of interfering with biological mechanisms of bacteria, they are focusing on the “efflux pumps” within bacterial walls. Efflux pumps are transport proteins that allow toxic substrates to leave the cell, ridding bacteria of harmful substances such as that caused by antibiotics taken into the cells. Efflux pumps have yet to be heavily influenced and pressured by evolution, so bacteria have similar types of these proteins.
"As a first in this field, we proposed the approach of essentially 'screwing up' the efflux pump's protein assembly, and this led to the discovery of molecules with a new type of antibacterial activity," said co-author Jeremy Smith, the UT-ORNL Governor's Chair and director of the UT-ORNL Center for Molecular Biophysics. "In contrast to previous approaches, our new mechanism uses mechanics to revive existing antibiotics' ability to fight infection."
Researchers combined chemical synthesis and culture experiments with supercomputer modeling to examine the function of efflux pumps and how they can be interrupted. Modeling on the ORNL’s supercomputer Titan allowed the group to simulate an extensive list of potential blockers before testing in the lab. The group scanned several potential chemicals to determine which ones would interfere with the protein assembly of the pumps. By simulating the dynamic nature of the proteins and testing how different chemicals interact with the changing protein structures, the list of potential drugs to test in the lab went from thousands to a handful. Four of potential chemical inhibitors were then synthesized and tested in bacteria to see whether they were able to interrupt the protein pump assembly. The study has been published in
ACS Infectious Diseases.
"The supercomputing power of Titan allowed us to perform large-scale simulations of the drug targets and to screen many potential compounds quickly," said Helen Zgurskaya, head of OU's Antibiotic Discovery and Resistance Group. "The information we received was combined with our experiments to select molecules that were found to work well, and this should drastically reduce the time needed to move from the experimental phase to clinical trials."
For this study, the team targeted a typical type of efflux pump found in
Escherichia coli bacteria. Since these efflux pumps tend to be similar across different kinds of bacteria, this approach could be expanded for testing on other Gram-negative bacteria. The team would like to perform further testing for additional potential inhibitor compounds and optimize the most potent combinations to work toward testing in clinical trials. More extensive simulations on the Titan supercomputer could also provide in-depth information about how the efflux pumps function and allow expanded testing based on this extended knowledge.
Sources:
phys.org,
ACS Infectious Disease,
Oxford Journals,
Farmers' Journal, Titan Image Courtesy of Oak Ridge National Laboratory, U.S. Dept. of Energy