Zoom-in On the Protein Behind Huntington's Disease
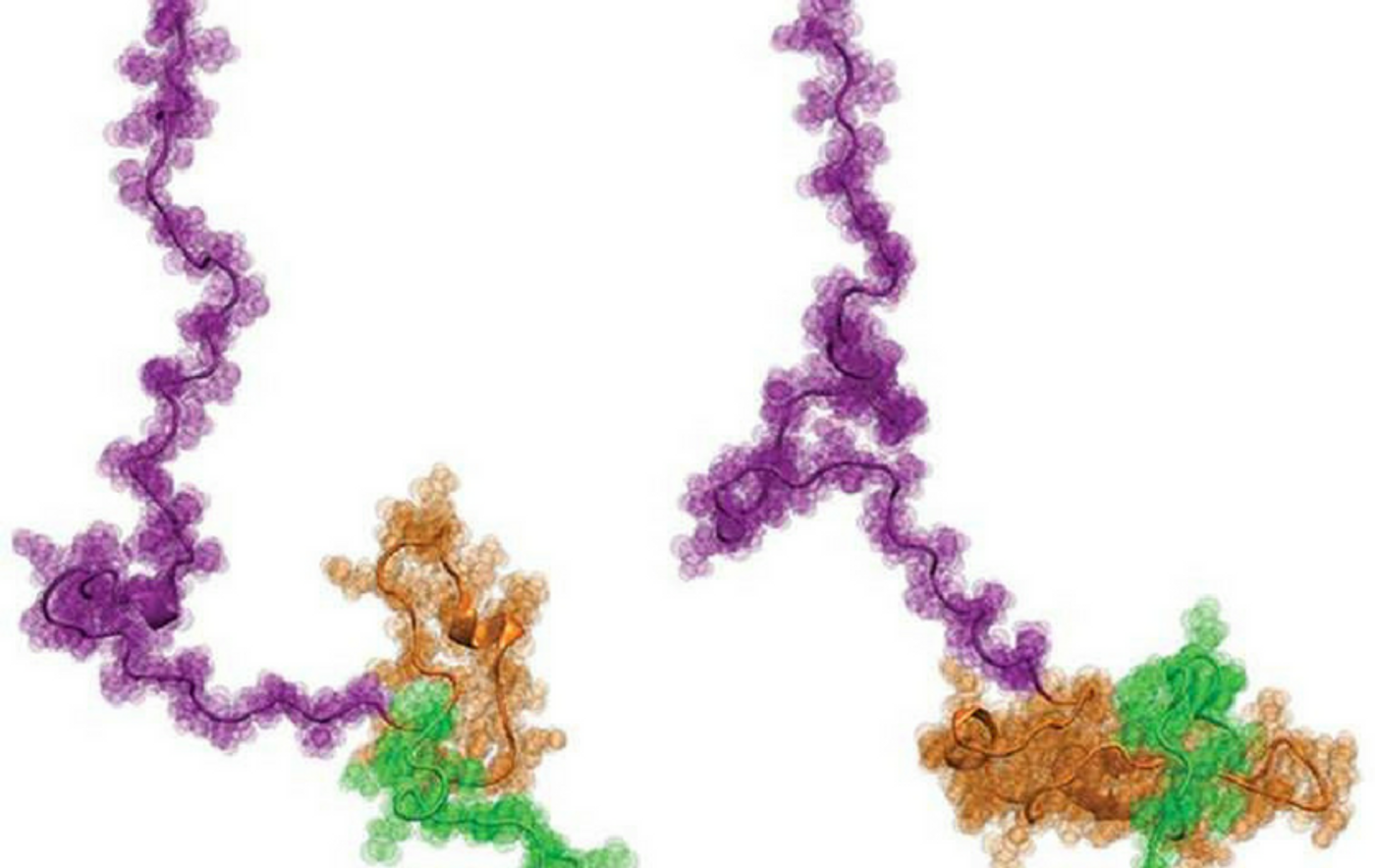
The structure of the huntingtin protein. Credit: Washington University in St. Louis
An international team of scientists and engineers have reported that they performed the first-ever atomic-resolution analysis on the protein behind Huntington's disease (HD) – huntingtin or Htt. By examining the structural feature of huntingtin closely, they hope that their data can help develop new therapies for this incurable disease.
Huntington's disease (HD) is a neurodegenerative disease caused by a mutated form of the huntingtin gene, where excessive trinucleotide CAG (cytosine-adenine-guanine) repeats result in the formation of an unstable protein. These expanded repeats get translated into a disease form huntingtin protein that contains an abnormally long polyglutamine (or polyQ) tract. The accumulation of this abnormal protein resulted in the death of brain cells. The early symptoms of HD are often subtle mood problems. As the disease advances, a patient’s body becomes uncoordinated, and movement gets jerky. The late stage of HD involves loss of mobility and vocal communication, as well as dementia.
There are only treatments to relieve some HD symptoms but no cure so far. Scientist believed that unraveling the structure of huntingtin protein in detail could lead to a cure eventually. But to obtain the atomic-level structural information of full-length protein and disease-relevant fragments (referred to as Httex1) have been proven challenging. As Rohit V. Pappu, an Engineering Professor at Washington University and a member of the international team, compared the process to “obtain the structural characterization of proteins within a mush”. The fragments have poor solubility, and the repeating nature of the polyQ generate an overlapping signal, making the structural studies somewhat tricky.
"Our goal was to gain insight into how increasing the length of the polyQ tail repeat alters structure of this protein at the monomer level and under conditions where we are able to unlink its folding and self-assembly," said Hilal A. Lashuel, a professor at the École Polytechnique fédérale de Lausanne (EPFL) and a collaborator of the project.
As a first step, precise, high-purity samples of Htt with site-specific fluorescent labels were generated and analyzed using molecular spectroscopy. For the second step of the project, single-molecule fluorescence resonance energy transfer (smFRET) was used to assess how the inter-atomic distances within Httex1 vary with the expansion mutations. (smFRET can measure distances between 1-10 nanometers within individual molecules). Data from the two steps were passed onto to Pappu's lab at Washington University. Pappu and his team developed novel computer modeling approaches to produce physically accurate, atomic-level structural models of Httex1.
The results from this international collaboration were quite a surprise. The overall structure of Httex1 resembles that of a tadpole, which contradicts the longstanding ideas about Httex1 accumulation in Huntington's disease.
Commenting on the tadpole-shape configuration of Httex1, Pappu said: “If the prevailing hypothesis were true, then the tadpole would have turned into a 'frog' as the polyQ length increases the threshold length, but that does not appear to be the case. The new results instead focus our attention on the novel gain-of-function cellular interactions that are driven by the tadpole structure with a larger polyQ head.”
The group of researchers is excited about the significance of their discovery. “While the prevailing hypothesis has favored a model where mutant huntingtin-induced toxicity is driven mainly by its propensity to misfold and aggregate, our findings suggest that aberrant interactions at the monomer level may also contribute to the initiation and/or progression of the disease,” explained Lashuel.
The next challenge for the scientists is to understand how these structural changes at the monomer level of Httex1 translate into increased aggregation and toxicity when the length of the polyQ tail crosses the pathogenic threshold.
Imaging a killer — Huntington’s disease. Credit: Washington University Engineering