Using Gene Networks to Get Closer to Personalized Medicine
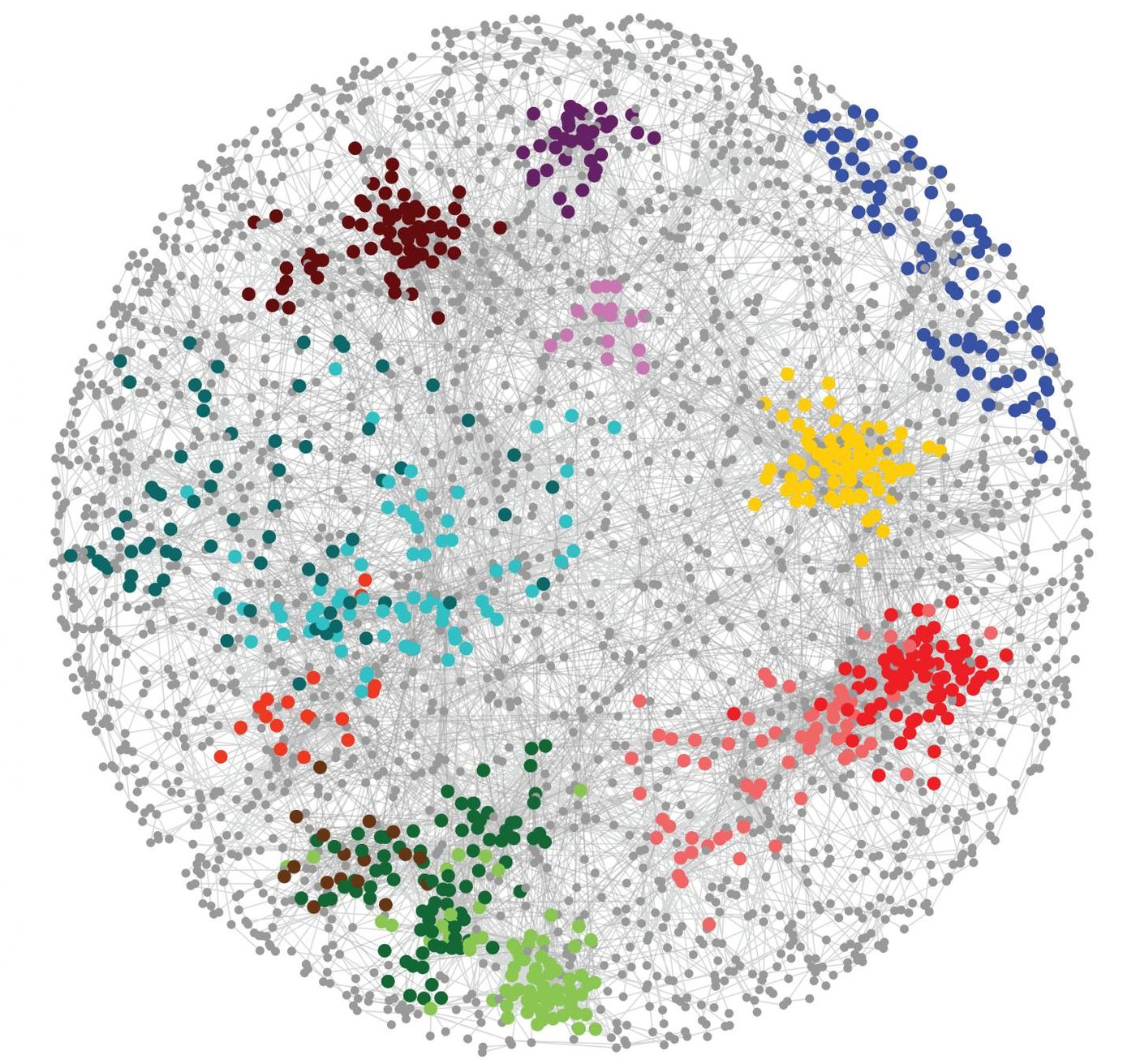
After a massive effort, the human genome was sequenced completely in 2003. Genetic sequencing technologies have been getting better and cheaper, and all of this knowledge has opened many doors. Now it’s not unusual for any random person to send a sample of their saliva off to a company that will then tell them more about their genetics, which has generated a ton of information. Scientists have to figure out how to handle all of this data and mine it for the knowledge we need. Researchers reporting in Cell have highlighted the importance of relating global networks of genes to health and disease instead of focusing on individual genes.
At one time, genes were studied largely by their dysfunction. People or animals that carried dysfunctional copies of genes had different biological characteristics. In some genes, a single error in one gene is enough to cause a severe disease. Over many years, scientists have learned a lot about such genetic diseases, and we now know what will happen to a person that carries a mutation in certain genes. But not all diseases that are caused by genetics trace back to a problem with a single gene. There may be many different genes, all carrying small changes, that pile up to increase, or sometimes decrease, a person’s risk of disease. Complex traits are affected by multiple genes, and now researchers are turning their attention toward these disorders, which can be much more difficult to understand.
"All the genome sequencing data is highlighting the complexity of inheritance for the human genetics community," said co-senior study author Brenda Andrews, University Professor and Director of the University of Toronto's Donnelly Centre for Cellular and Biomolecular Research. "The simple idea of a single gene leading to a single disease is more likely to be an exception than a rule," she said.
GWAS, or Genome-wide association studies are large investigations that compile the genetic data of many people and identify small changes in genes, called variants, that are connected to some characteristic(they are outlined in the video below). Many of those variants have a tiny effect, so it can be hard to find them unless one looks at huge numbers of people. Other gene variants have a more powerful impact or one that gets stronger when there is some environmental condition present. Complicating it even further, some variations can have a positive effect.
"It would be a simpler problem if one particular mutation resulted in Disease X all of the time, but that's often not the case," explained study author Michael Costanzo, Senior Research Associate in Boone's lab. "To understand the effect of combinations of variants is really difficult. We suspect it's particular sets of mutations that really impact what the disease outcome is going to be in a personal genome. How genes interact with each other is important and, given our current understanding of gene-gene interactions, it's not a problem that's easily solved by reading individual genome sequences."
To illustrate the challenges in analyzing genomic variants, consider this calculation by scientists at the Broad Institute of Harvard and MIT: identifying a gene pair that underlies a disease would require genomic sequences from half a million patients with the disease, and an additional half million unaffected individuals as controls. "If most genetic diseases involve gene combinations, collecting enough patient data to find these interactions is a huge challenge," noted Costanzo.
The function of a gene can be predicted by studying a simple model organism like yeast, and networks of genes can be constructed with this data. If two genes interact, it means that the effect of both at once is greater than what would be expected from each on their own. Modifier genes could also exert an impact; they reduce the impact of damaging mutations. While it’s a relatively straightforward process to tinker with gene expression in model organisms, it’s obviously much harder to test these effects in people.
The CRISPR gene editing tool can now create cell lines with known variants and introduce new ones. This helps show how combinations of genes can influence human disease, which can now also be combined with the massive amount of new data from human health and genomic databases.
Knowing more about genetic interactions and gene networks can not only help researchers know more about health and disease and lead to more personalized medicine. It can also help reveal new drug targets or therapeutic routes and can demonstrate why drugs can have different effects on different people. The work will also apply to cancer research; cancer is a disease caused by genetic problems. Scientists can now study how genes change in cancerous cells, and potentially, develop drugs that target cancer mutations specifically. The knowledge of genetic interactions will also help shed light on why so many approved cancer drugs only work in some patients and not others.
"We can't think about genes in isolation anymore," said co-senior study author Professor Charles Boone. "We have to start looking at variants of multiple genes as a major component of genetic disease, because those combinations are going to be different for different people and these specific combinations may not only profoundly affect disease susceptibility, but they will likely dictate new personalized therapies."
Sources: AAAS/Eurekalert! via University of Toronto, Cell