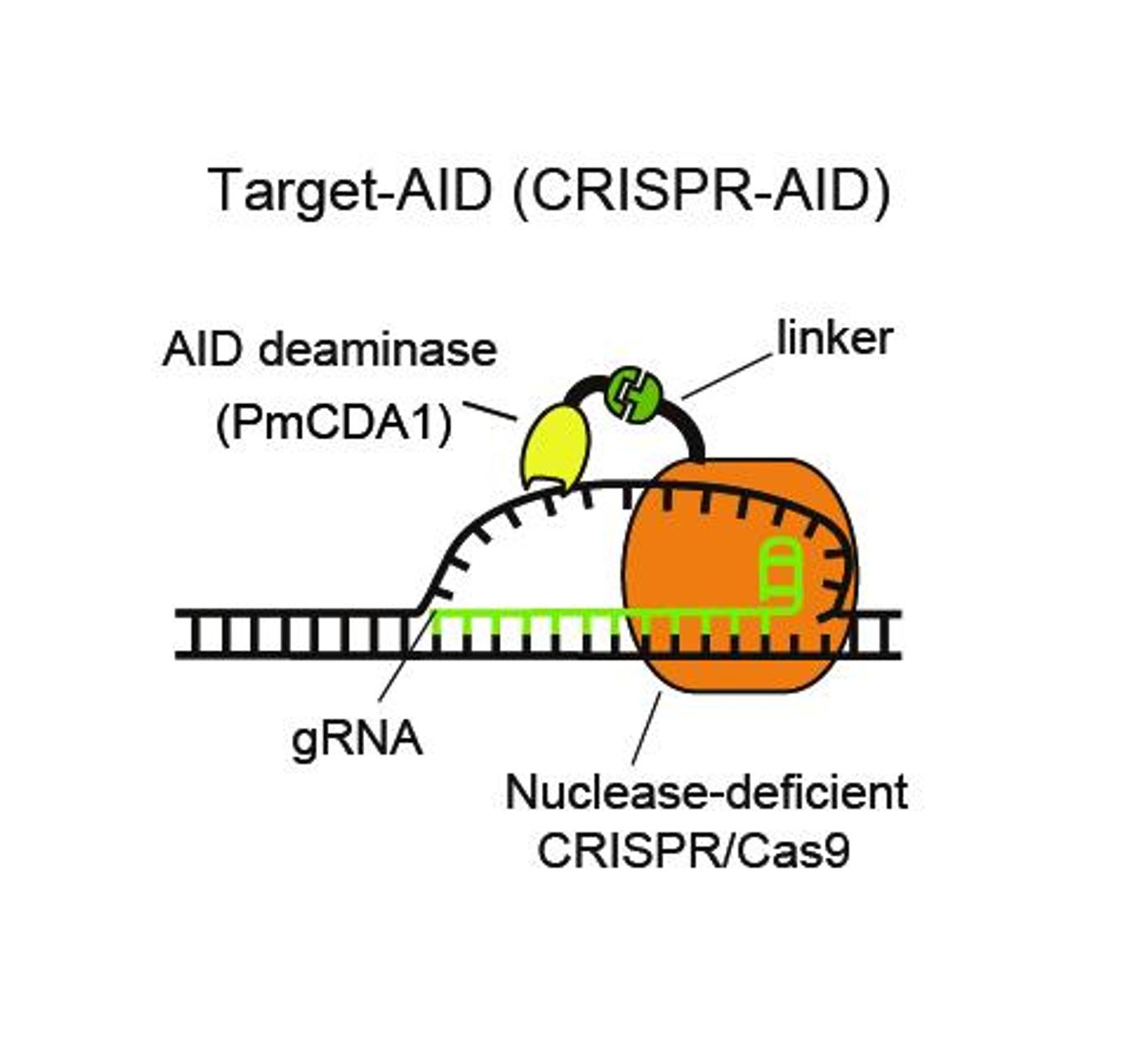
Researchers have developed a genome editing technique that does not require DNA to be cleaved. Called Target-AID, the technique builds on the CRISPR/Cas9 tool but aims to avoid the toxic effects that can result from cutting through genomic DNA. The work was done by scientists at Kobe University in Japan and
published in Science.
The revolution in genomics set off by the development of CRISPR/Cas9 technology has been unprecedented. In bacteria, CRISPR/Cas9 functions as an immune system that fights foreign invaders by destroying their DNA. In the lab, the function was harnessed so the bacterial nuclease enzyme cuts DNA at a site specified by the researcher. The targeted genetic material is then altered when the cut is repaired by cellular machinery. For a detailed description of that technology, check out the video below.
The new technique seeks to avoid a caveat of the CRISPR/Cas9 technology – unintended detrimental effects from cutting chromosomes. The researchers engineered a CRISPR/Cas9 system that did not cut DNA, and combined it with AID. AID (Activation-induced cytidine deaminase) can mutate DNA by altering the chemistry of the cytosine nucleotide base – by deaminating it - to change it to a uracil. The C:G base pair in the genomic DNA becomes a mismatched U:G. Cellular machinery will see the uracil as a thymine, so during replication what was a C:G becomes a T:A base pair.
“Nuclease-deficient type II CRISPR/Cas9 and the activation-induced cytidine deaminase (AID) ortholog PmCDA1 were engineered to form a synthetic complex (Target-AID) that performs highly efficient target-specific mutagenesis,” the researchers explained in their publication. “Specific point mutation was induced majorly at cytidines within the target range of five bases.
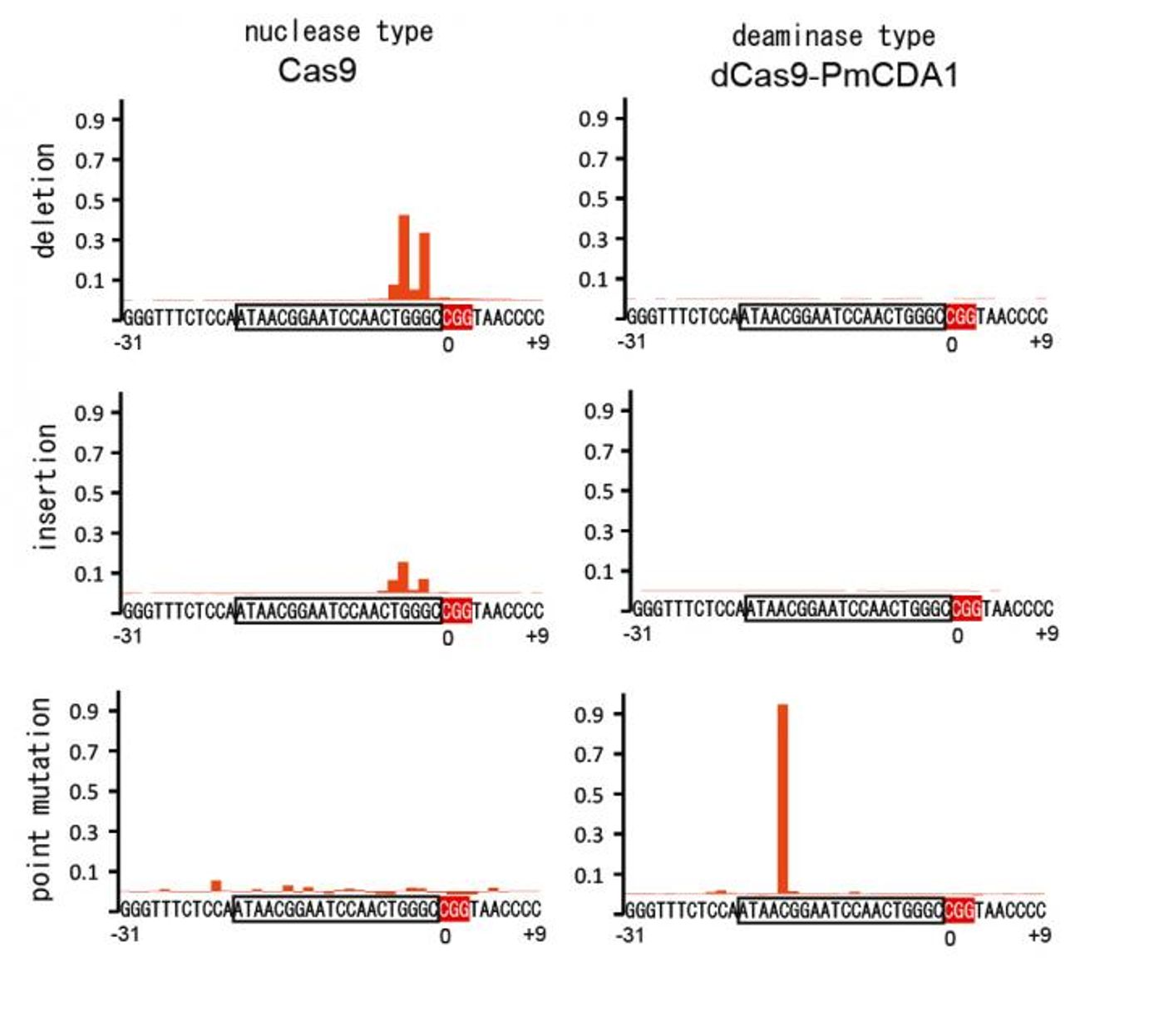
The researchers demonstrate in their paper that this is an efficient and effective method for creating targeted point mutations. They also show that compared to other nuclease methods, cytotoxicity or cell death is greatly reduced.
It’s also suggested that other deaminases could be used in place of the one utilized by the scientists in this study. The deaminase used in this work was derived from a lamprey. Alternatively, the rat cytidine deaminase might be a suitable substitute.
“Although direct comparison will be needed, these two systems may complement each other to extend the repertoire of possible editing sites,” write the authors in the paper. “Using other CRISPR-related systems or other modifier enzymes such as adenosine deaminase also will broaden the editing capacity and further enrich the genome editing toolbox.”
Notably, the investigators found that the combination of deaminase with Cas9(D10A) was effective in yeast and caused insertions and deletions (indel) in mammalian cells. When a uracil DNA glycosylase inhibitor was introduced, the indel formation was reduced and efficiency increased.
If you'd like to know more about activation-induced cytidine deaminases, watch the brief video above that shows the dynamic mechanism of deamination, which occurs so that antibody diversity is generated in the immune system.
Sources:
AAAS/Eurekalert! via
Kobe University,
Science