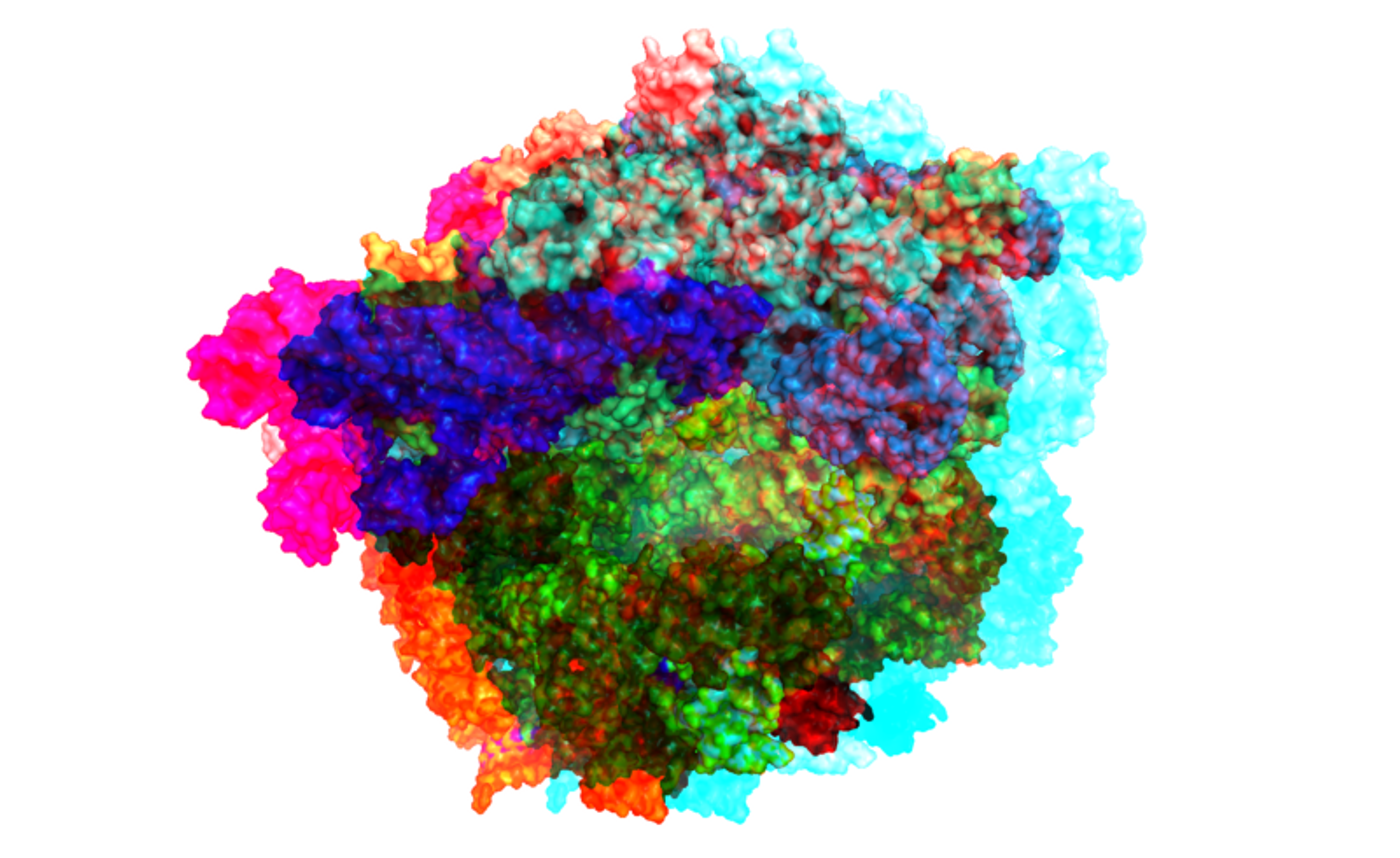
Our DNA uses messenger RNA or mRNA as the intermediary that directs cellular machinery to construct the many various proteins that compose our body. Cells must also know when to stop making protein and maintain the right levels, something that is vital to health and cellular function. New work led by Jeff Coller, PhD, the Director of the Center for RNA Molecular Biology at Case Western Reserve University School of Medicine, has revealed the mechanism that keeps that balance in place.
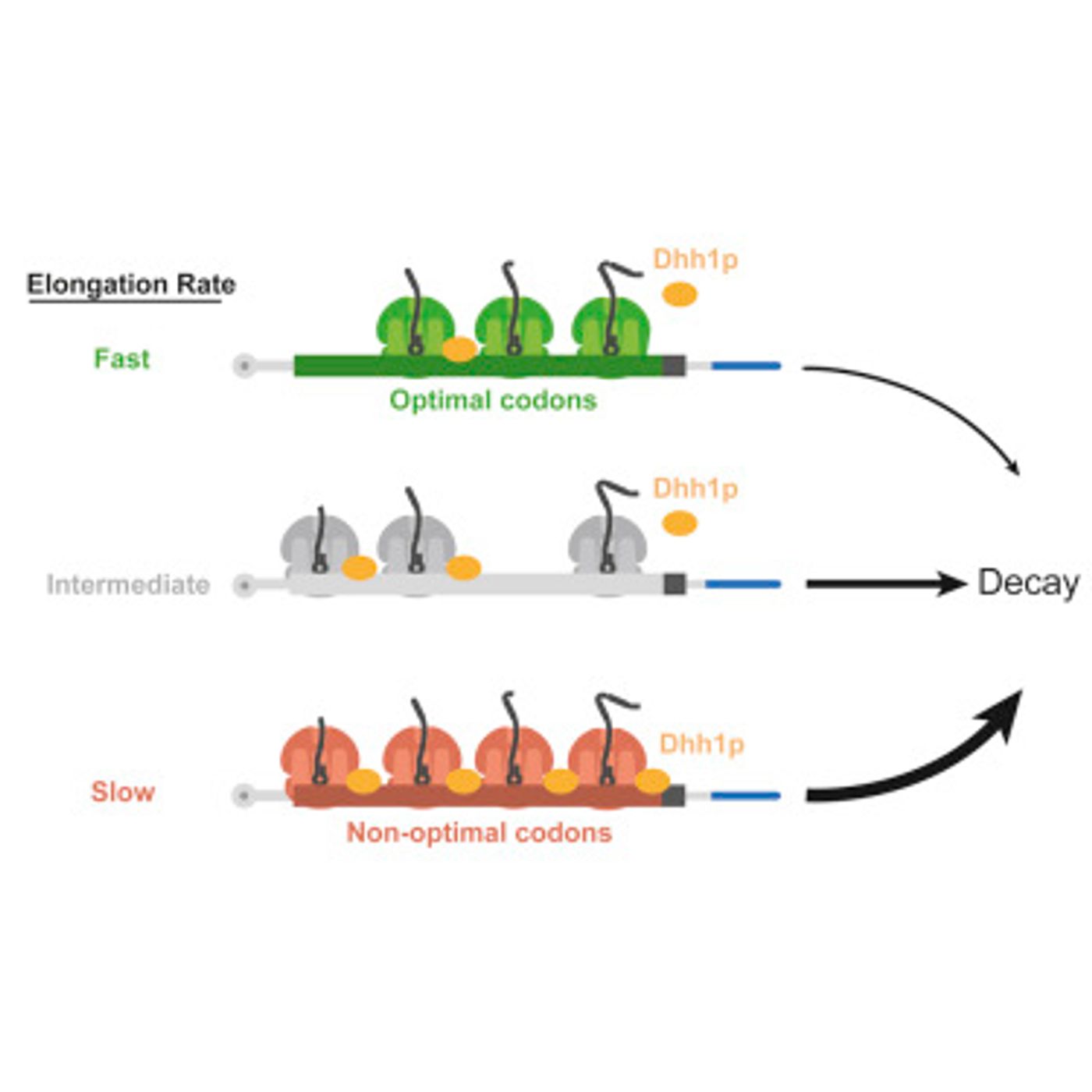
The study, published in Cell, illustrates that an enzyme called DEAD-box protein Dhh1p can determine if mRNA molecules are needed, or if they have to be eliminated. When cells require a protein, mRNA is made from the gene that encodes for it. After the cell has a sufficient amount of the protein, the corresponding mRNA is removed. That is a complicated process in which enzymes recognize portions or tags on the mRNA that indicate that it’s meant to be destroyed. Coller’s research team has determined that Dhh1p can target mRNA for destruction when the mRNAs are being translated into protein too slowly.
"We knew there had to be some way to communicate how fast the genetic code was read to the machinery that ensures the message is removed from the cell," commented Coller. "There had to be a way to put on the brakes."
According to the study, "Dhh1p is a sensor of slow ribosomes and communicates this information to the mRNA decay machinery." The work indicates that genetic sequences are an important factor in mRNA stability, and a sequence change as small as 10 percent can influence the ultimate fate of an mRNA molecule.
Ribosomes can translate common mRNA sequences quickly, while less abundant sequences get translated more slowly. The researchers found that Dhh1p binds to mRNA with sequences that are not optimal and can physically interact with ribosomes that are moving slowly.
This study has revealed important details about the regulation of protein levels inside of cells. The knowledge may give insight into therapeutic development that tinker with this process.
"Our study provides a new way to look at the genetic code," explained Coller. "We're so used to looking at how DNA mutations cause a change in protein function. We must also consider how enzymes like Dhh1p sense the speed at which ribosomes interpret the genetic code. Now I can look at the genetic code in terms of speed and rate, and with reasonable accuracy predict how much protein is going to come from a gene. There's huge application for that in human biologics, proteins that are easily taken by injection."
"There are rare genetic diseases attributed to RNA being read too slow or too fast. We can now manipulate this process to dial up or down protein expression. The speed at which the ribosome reads the genetic code and is sensed by Dhh1p could open up a new set of mutation types that could indicate disease states we are unaware of today," Coller concluded.
If you would like to know more about protein synthesis, check out the lecture above from Rachel Green of Johns Hopkins University and Howard Hughes Medical Institute.
Sources:
Phys.org via
Case Western Reserve University,
Cell