Decoy sequences of DNA are able to interfere with gene expression, and in ways that are not yet well understood. New findings by researchers at Rice University, who used computer simulations to help them understand how complex molecular pathways are regulated in cells, have implications for the design of drug treatments.
The work was published in the journal Interface by lead authors Zhipeng Wang and Davit Potoyan who worked with biophysicist Poeter Wolynes. The takeaway message of the research, said Wolynes, is that instead of focusing only on certain molecular targets, manufacturers must investigate how drug products impact complete biological networks.
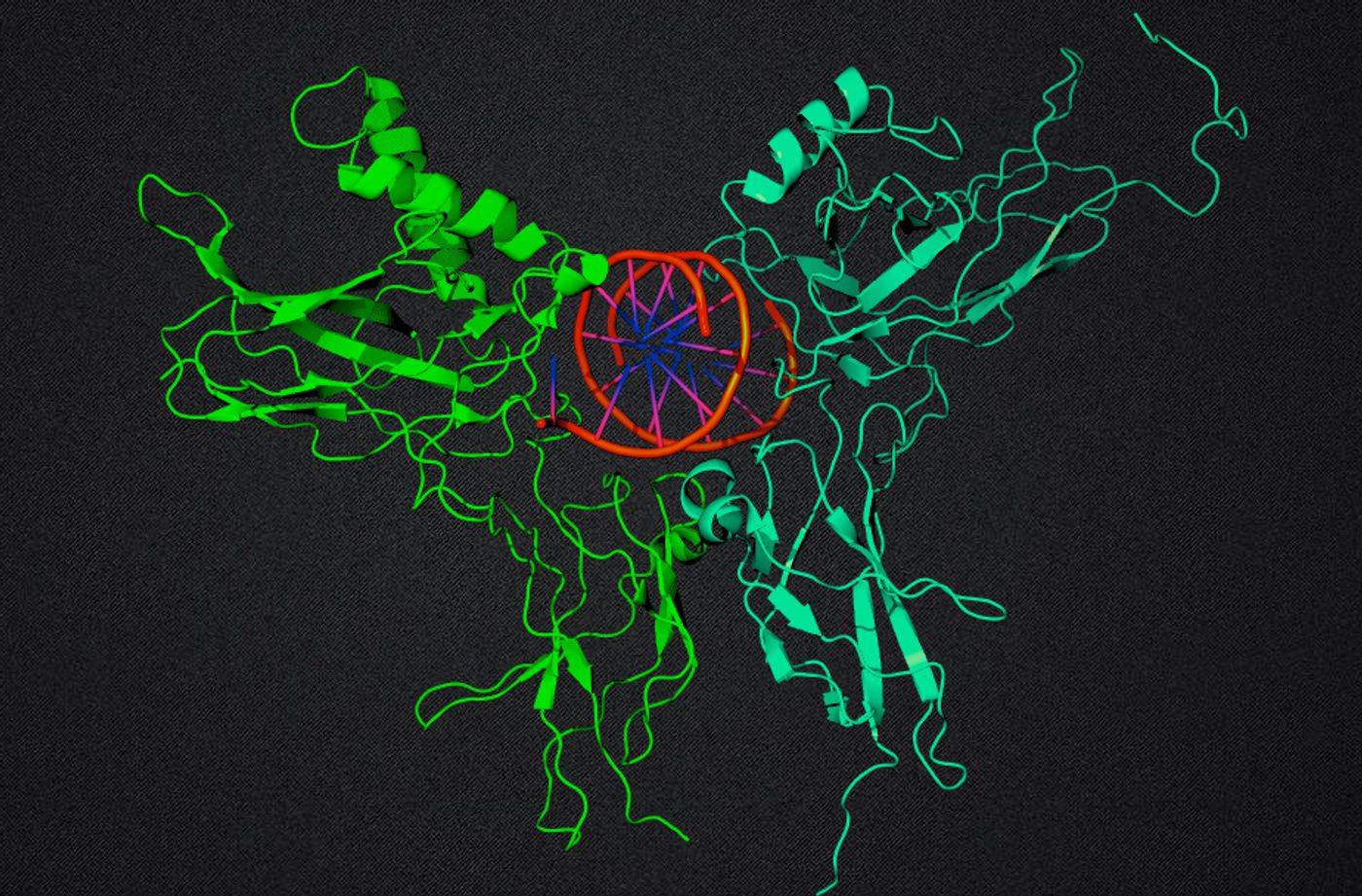
Transcription factors bind to DNA and exert an influence on gene expression. In this study, the researchers were focused on a network of cellular pathways that is affected by a family of transcription factors called NFkB. Dysfunction in the regulation of NFkB has been implicated in many diseases including cancer, arthritis, heart disease, and asthma.
NFkB works in conjunction with the enzyme complex IkB; gene expression is modulated to control events like cellular death, growth and differentiation, and the immune response to inflammation. Normally, NFkB binds to DNA, which prompts IkBa expression. However, some NFkB molecules bind to decoy DNA sequences instead of to functional targets. A recently discovered process called stripping then occurs in which all of the bound NFkB are released at once from the DNA, which stops transcription. This cycle of NFkB binding and unbinding is critical to the expression of many different proteins, but the importance of the seemingly non-functional decoy sites is not understood.
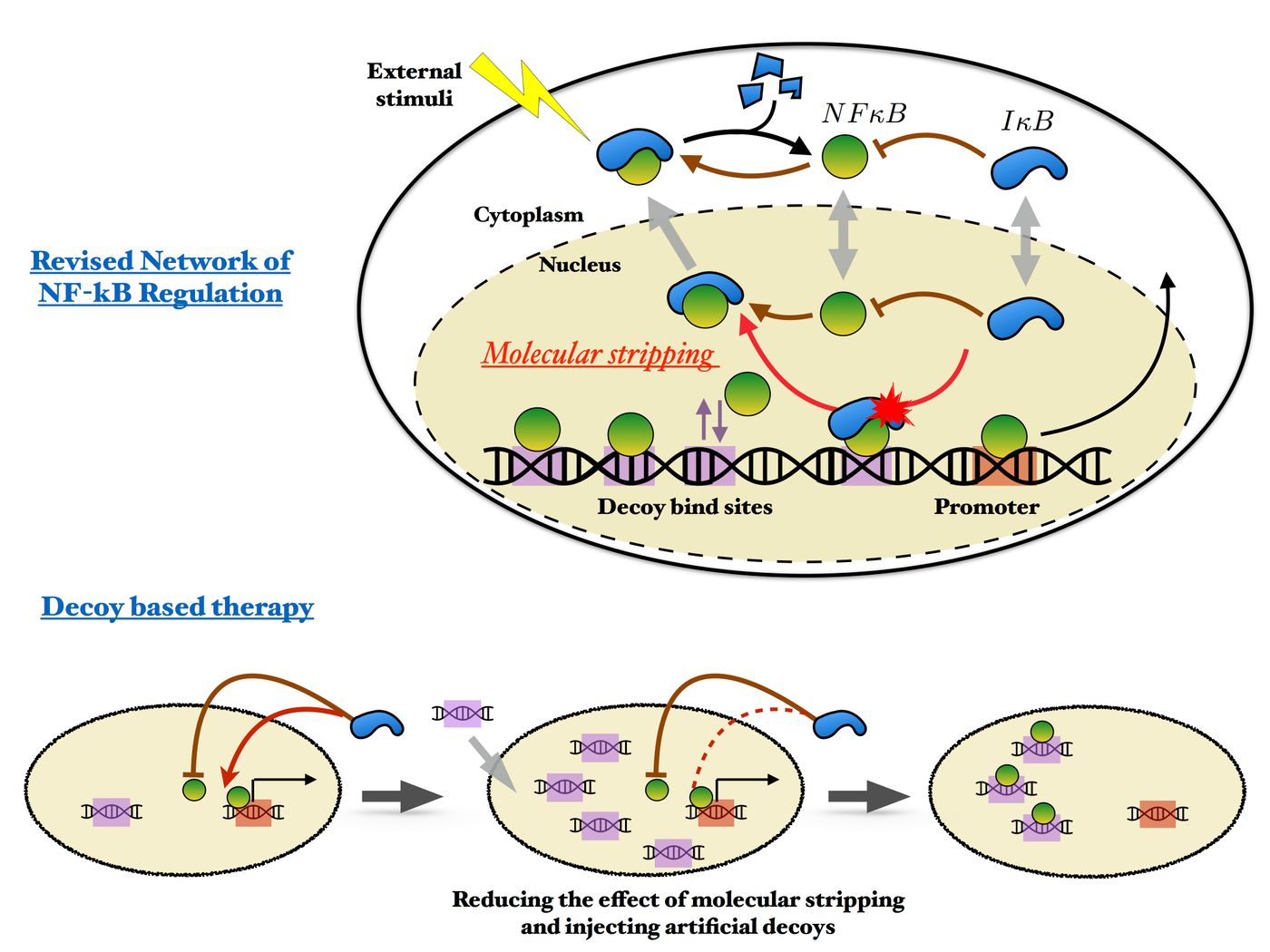
“Some transcription factors can bind to many sites, and sometimes this binding is nonspecific, with no apparent purpose. Traditionally, people look for things that change dramatically,” explained Potoyan, who is a postdoctoral researcher in Wolynes’ lab. “They look for something that turns on, something synthesizing, something degrading. Decoy binding doesn’t appear to be very interesting, but we’re saying it can influence system-level dynamics in a quiet way by sequestering transcription factors from being active in the nucleus.
“In this case, we illustrate the decoy problem with the NFkB example showing how decoys can change the frequency and modulate the amplitude of the circuit.”
Wolynes explained that IkB strips proteins both from all of the DNA binding sites without discretion. “One consequence is that it becomes harder to recognize an active site from a decoy, throwing a considerable monkey wrench into the bioinformatics problem of how to find interesting parts of the genome.”
Synthetic sequences of DNA have been used as drugs to try to control gene expression to a patient’s advantage, including attempts to treat NFkB misregulation. “But I don’t think these treatments are yet as successful as they could be,” Wolynes said. “This paper shows the effect of the decoys will be diminished by stripping. One of the ‘translational’ suggestions in this paper is you’ve also got to control the level of IkB to use decoys more effectively, and that suggests some kind of combination therapy.”
Wang, a graduate student at Rice, would like to see the team’s increasingly comprehensive models of gene networks used to aid drug designers. “I want to use physical chemistry and physics principles to provide quantitative guidance to translational researchers and physicians so they can better understand how to cure disease,” he said.
Source:
Rice University,
Interface