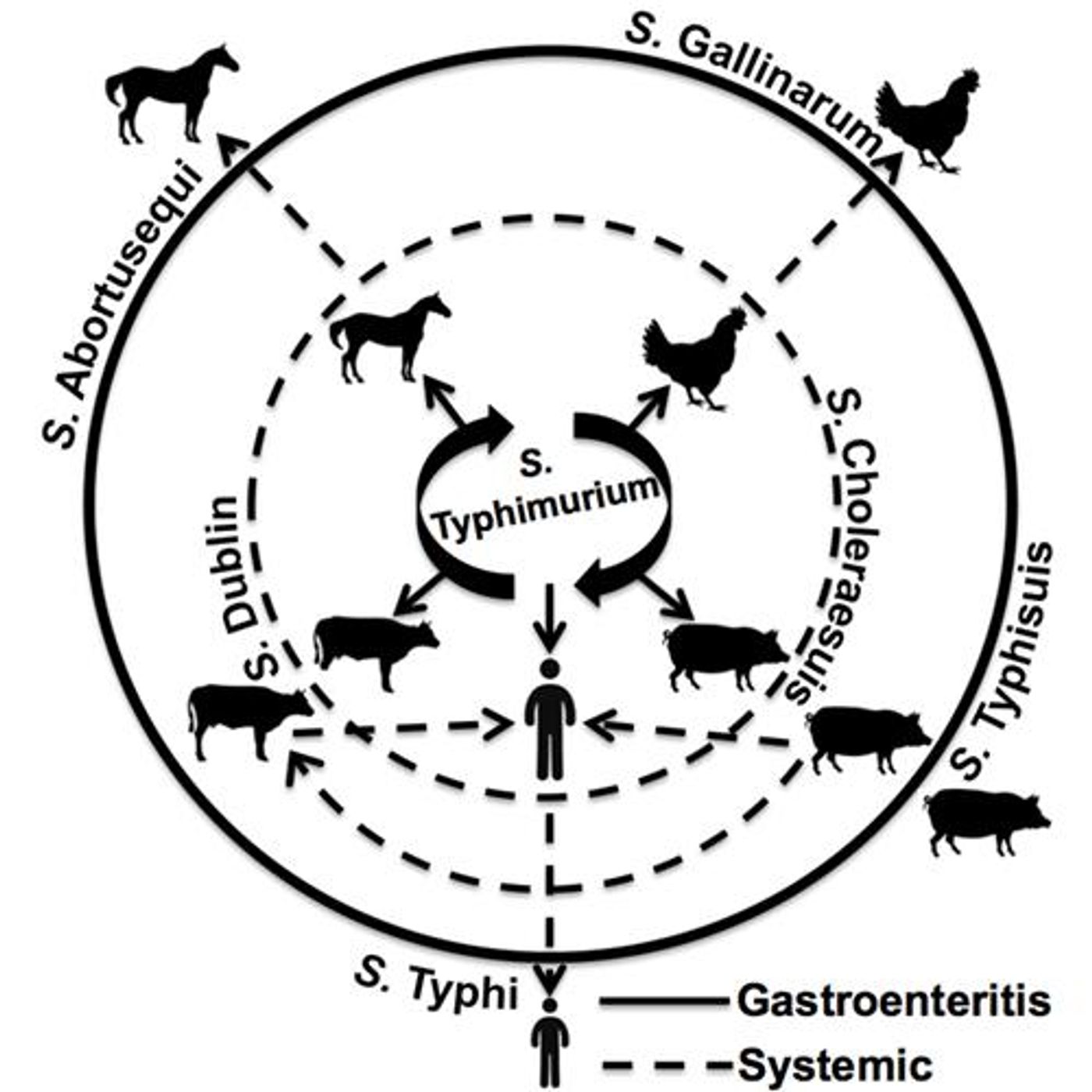
What makes a strain of
Salmonella infect cows, but not humans or birds?
Researchers at the University of Pennsylvania combed the genomes of 580
Salmonella strains for mutations that could determine host specificity. The group used a technique called “
genome-wide association studies” (GWASs) to identify the mutations. GWASs analyze genome sequences to identify mutations associated with specific traits (like diseases). Often, these analyses focus on SNPs -
single nucleotide polymorphisms. SNPs are genetic mutations that change one DNA base to another - an adenine into a thymine, for example. Many SNPs are harmless, but some result in amino acid changes.
Salmonella enterica serovar Typhimurium contained a large number of SNPs within genes for surface and secreted proteins. This particular serovar (or strain) is a common cause of food poisoning in humans. Humans contract this strain of
Salmonella by eating contaminated food. Since
Salmonella is a facultative intracellular pathogen, it is able to infect cells that line the intestine, leading to symptoms of food poisoning (diarrhea, nausea, vomiting).
Salmonella prefers to live in association with its host, but is able to survive in the environment - on produce, for example - for weeks.
According to study author Dieter Schifferli, “we saw this huge variation in proteins on the surface of bacteria or in secretions, which are really the first lines of interaction with the host … if there was so much variation, it suggests it must be linked to something important”.
Based on their findings, the group focused their attention on the gene for the proteinaceous surface adhesin FimH. They introduced cow and human-specific fimH mutations into
E. coli and tested the bacteria’s ability to adhere to host cells. They found that a single amino acid change in human-specific FimH caused
E. coli to preferentially bind to cow cells. In other words, this relatively small mutation was responsible for a change in host specificity.
Schifferli now plans to determine whether there are genetic differences that determine whether
Salmonella will cause mild food poisoning or more serious disease.
Sources:
Eurekalert,
Salmonella.org, Wikipedia