A bacterium called Burkholderia multivorans adapts and changes in bursts in order to survive in the lungs of cystic fibrosis (CF) patients, according to a report published this week in
mSystems, an open access journal from the American Society for Microbiology.
CF patients worldwide typically get chronic airway infections from Burkholderia bacteria; B. multivorans is the species most commonly isolated from those infections. While sometimes the patients just expel it, in most other cases it becomes a serious infection that persistently infects the lungs for many years. Study author Leonilde M. Moreira, PhD, who is an assistant professor at the Instituto Superior Técnico in Lisbon, Portugal, says that "... our understanding of the traits required for bacterial colonization and persistence, as well as the molecular mechanisms underlying this adaptation, are limited."
To better understand this evolution, Moreira and colleagues examined phlegm specimens from a CF patient over a period of 20 years. This particular patient was hospitalized once for a lung illness related to CF as well as having received multiple courses of antibiotics over that time period.
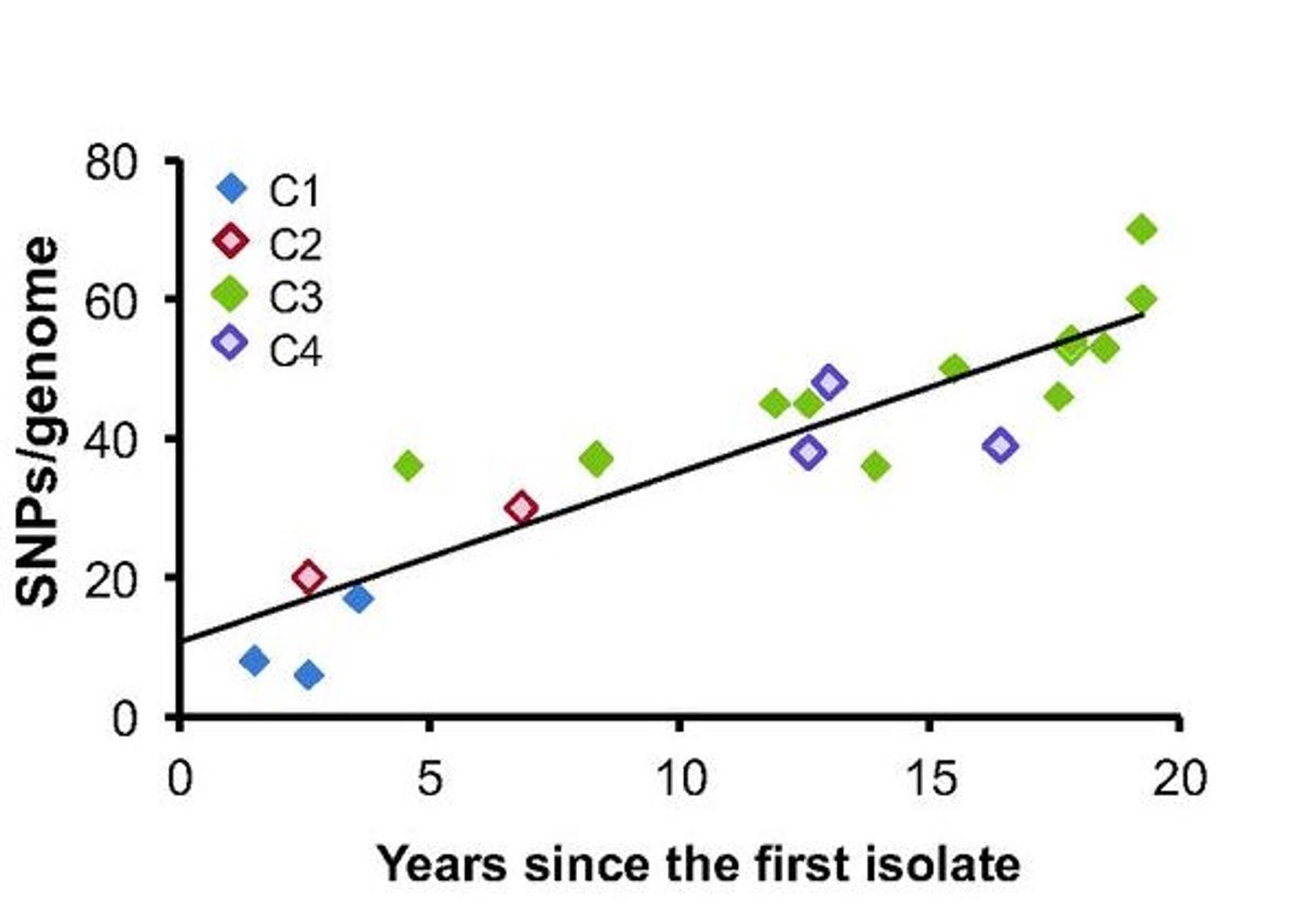
Mooreira’s team wanted to analyze genomic and functional alterations by sequencing the genomes of 22 isolates of B. multivorans taken from the patient, then comparing the results with the patient's medical records to look for correlations. They found that several unique bacterial lineages coexisted at any given time but were evolving at different rates. These new lineages changed primarily with mutations in genes with regulatory or signaling roles, and in genes whose proteins are involved in metabolism. One family had rapidly diversified into three others. They observed a slow, steady and population-wide rate of genetic change occurring over the course of infection, at a rate of roughly two single nucleotide polymorphisms per year.
"These mutations corresponded to what was happening physically with the patient, so we could see that those mutations were not just random -- they were specific targets that affected the physiology of the bacteria." It is implied based on the evidence that the modifications the researchers observed were allowing the bacteria to survive.
A sample of B. multivorans was first taken from the patient in 1993; more samples were isolated periodically until 2013. She was first infected by Staphylococcus aureus, Haemophilus influenzae and Pseudomonas aeruginosa in 1989-1993. A period of co-infection of these three microorganisms with B. multivorans occurred from 1993-1996. The evolution was characterized by stages of strong diversification followed by intervals of relative stability. The time of highest diversification within the B. multivorans infection was correlated with more rapid deterioration of lung function in the patient, said Moreira.
"Altogether, our observations suggest that B. multivorans populations, during long-term colonization of the CF lungs, either directly or indirectly target adherence, metabolism, and changes in the cell envelope related to adaptation," Moreira said. “This dynamic suggests that monitoring these evolutionary and molecular patterns could be used to design responsive therapies designed to limit population diversity and disease progression."
The study unfortunately analyzed only a single patient, so these results must be repeated, Moreira said. Her team now continues the study of the bacteria in an additional 10 CF patients.
Sources:
AAAS,
mSystems